The latest VarSeq release offers enhanced control over genome browse visualization computations. These computations, which are responsible for computing and caching coverage files that create the aggregate view of tracks within GenomeBrowse, are now more flexible. Typically, these tracks create files with the “.covtsf” extension and are used for zoomed-out density views of sources like VCFs and BAMs.
Managing Automatic Track Computation in GenomeBrowse
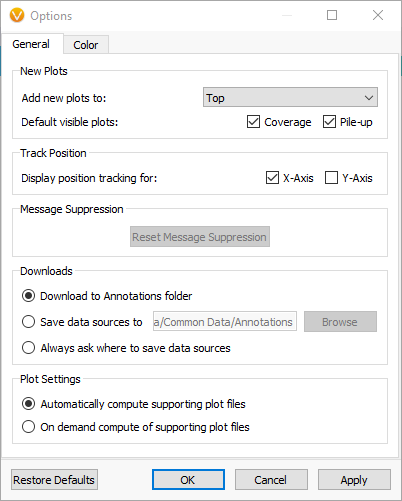
Previously, the computation would commence automatically when you plotted a source in GenomeBrowse. While you could always cancel it by clicking the “X” on the progress bar in the top-left corner, you can now customize the default computation behavior. To access these settings, navigate to the gear menu in the GenomeBrowse toolbar and click to open the options dialog.
The plot auto-computation settings can be found at the bottom of the general tab. By default, it’s configured to automatically compute supporting plot files. However, you now have the option to compute them only on demand. With this setting, if you plot a file lacking computation files, you’ll see a button at the bottom of the plot that allows you to initiate the computation when needed.
It’s worth noting that you can still view these tracks in a zoomed-in mode. If you’re navigating between variants, you may not always need to compute the aggregate coverage tracks, saving valuable background computation time.
Should you have any questions regarding these settings and their impact on your workflow, don’t hesitate to contact our Field Application Scientist (FAS) team at [email protected].
Want to get in touch with our team to learn more about our comprehensive software suite? Please contact them today at [email protected].