Ever since the MacArther Lab announced the new gnomAD browser at last year’s ASHG conference, we have had many requests from our customers to make this new variant frequency source available within both VarSeq and SVS.
This new dataset includes variants obtained from 123,136 exome sequences and 15,496 whole-genome sequences. In comparison to the original ExAC dataset which contained exomes from approximately 60K samples, the 1kG dataset of about 2,500 and the ESP Exome dataset of about 6,500, this is by far the largest sample source of variant frequencies publicly available.
In addition to an increased number of individuals for each of the original ExAC populations the dataset now includes allele frequencies across over 5,000 Ashkenazi Jewish (ASJ) individuals. See the release notes for further specifics.
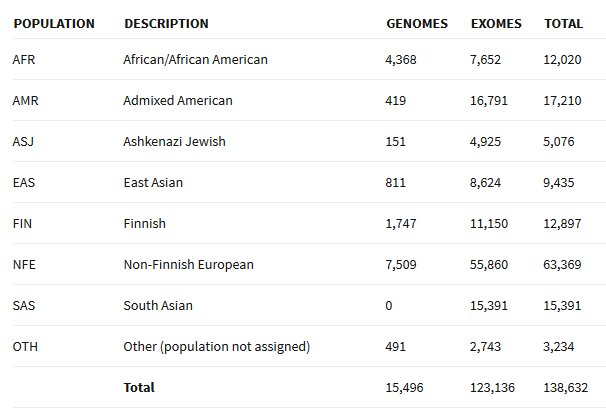
Figure 1. Break down of population groups as provided from the release announcement.
We will be curating the data into two annotation sources, one for the frequencies from the Exomes samples and one for the frequencies from the Genomes samples. Both new tracks will be made available alongside the original ExAC annotation source for use in any Golden Helix product.
For these new sources, we have minimized the unnecessary fields to save on the size of the track. The new Exomes track contains more variants than the original ExAC source (over 17 million in the new source compared to about 10 million variants in the original ExAC) with an extra population for nearly the same size file.
The filter field provided in these new sources is an important field to reference when using these annotation sources (see recent blog done by Gabe Rudy for a case study). Any variant with a flag other than PASS should be considered suspect. Filtering is now done on a per-allele level and one of the most useful filters is the new “Random Forest (RF)” filter which does some machine learning prediction on low-quality variants. Further details on each filter option is available in the release blog.
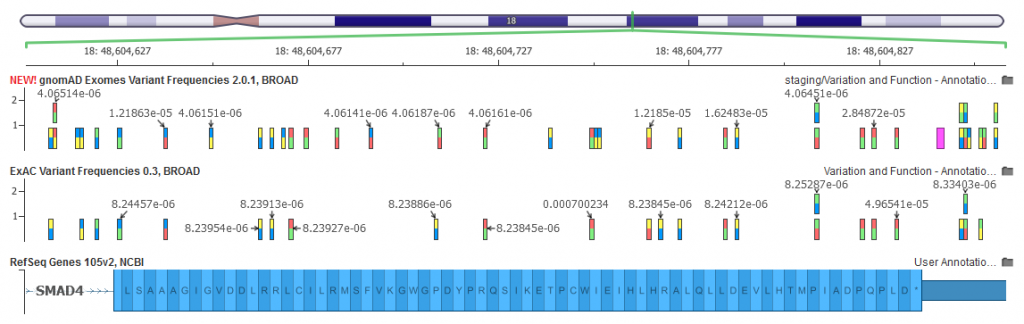
Figure 2. Exon 12 of the SMAD4 Gene showing the additional variant density for gnomAD.
Keep an eye out in the next few days for the Exomes annotation source with the genomes source following a few days later. Email us at [email protected] if you have any questions about this or any other annotation sources.
I would like clarify if the population for the exomes in the gnomAD is completely different from the ExAC population.
Hi Caline,
Think of the new gnomAD tracks as expansions of the original ExAC database. This blog post from the MacArthur Lab describes gnomAD tracks in great detail. The gnomAD tracks contain more variants than the original ExAC source (over 17 million in the new source compared to about 10 million variants in the original ExAC track.