Learn about the latest cancer ontology software updates and how they can enhance your research and work.
Cancer Ontology
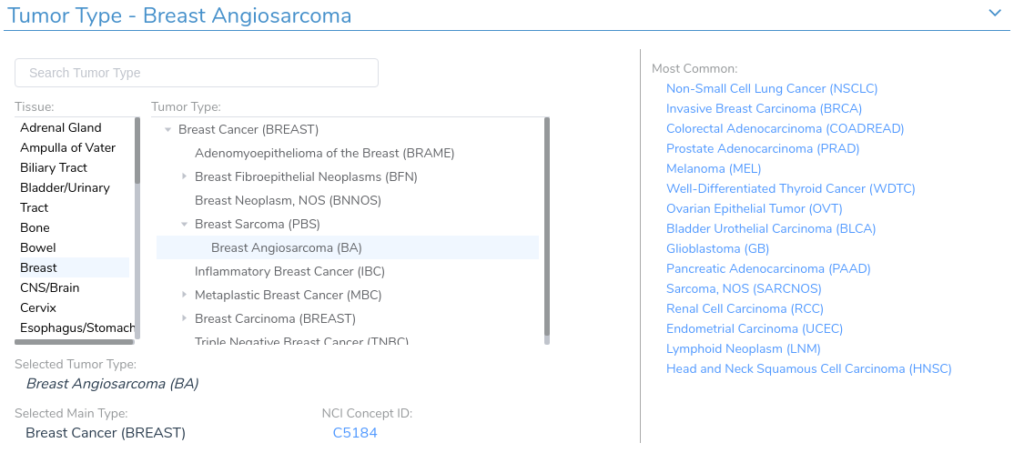
An important feature of the VSClinical AMP workflow is the ability to select the correct tumor type for a given evaluation. The selected tumor type is used to identify relevant clinical evidence, including drug sensitivities, resistances, and previous biomarker interpretations.
This functionality is powered by the VarSeq Cancer Ontology, which is a tree-structured cancer classification system in which root nodes represent general tissue types and branches represent different tumor type sub-classifications with increasing specificity as you move toward the leaves of the tree.
The VarSeq Cancer Ontology was inspired by the OncoTree [1] cancer classification system developed by Kundra et al . While much of the structure of our ontology was initially based on OncoTree, we have deviated from this system significantly since the initial release of the VSClinical AMP workflow and, with the help of our excellent CancerKB curation team, we have completely restructured the sub-trees for a number of tissue-types to better meet the needs of our users.
Breast Cancer Sub-Tree
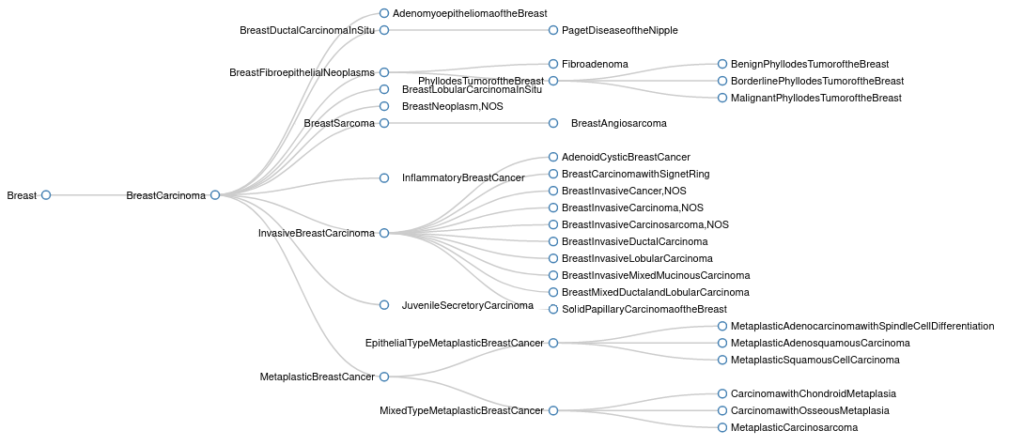
Perhaps the best example of this restructuring can be seen in the breast cancer sub-tree. The previous version of this sub-tree is shown below.
There are a number of issues with the structure of this tree. First, the root tumor type is Breast Carcinoma, which is not technically correct, as there are several breast cancer sub-types that should not be classified as carcinomas. Second, Triple Negative Breast Cancer is missing from the ontology entirely, despite being a commonly used classification to distinguish breast cancers which are negative for expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Third, Juvenile Secretory Carcinoma of the Breast is no longer considered to be the preferred nomenclature, as it is now known that this type of carcinoma can develop in any age group. These issues have been resolved with the updated version of the breast cancer sub-tree, which is shown below.
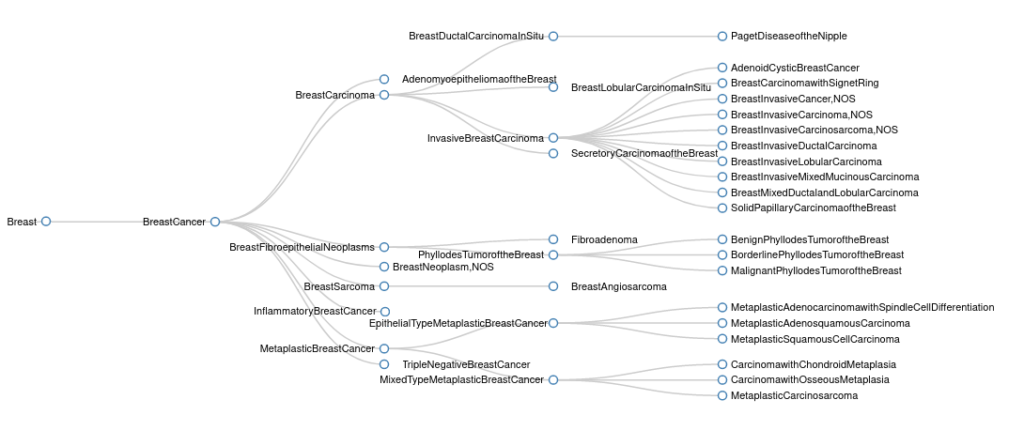
In the updated sub-tree, the root tumor type now has the more general name of Breast Cancer and a separate Breast Carcinoma sub-tree has been created. All forms of breast carcinoma are now grouped under this sub-tree, and all forms of breast cancer that are not classified as carcinoma now have their own distinct branches under the more general Breast Cancer tumor type. We have also added a new Triple Negative Breast Cancer tumor type and renamed Juvenile Secretory Carcinoma to Secretory Carcinoma of the Breast.
Conclusion
This is just a brief highlight of the improvements that we have made to the Cancer Ontology over the past year, and similar restructuring has been performed to improve the Lung, Cervix, Esophagus/Stomach, and Bladder sub-trees as well. While we take inspiration from existing ontologies, such as OncoTree and the NCI Thesaurus [2], none of these existing ontologies provided a perfect solution to our needs. This has led us to develop our own independent Cancer Ontology to provide a robust hierarchy of the most common tumor types for the interpretation of sequence variants in cancer. We will continue making improvements to our ontology, and new versions will be made available periodically, which can be downloaded using the VarSeq data source manager. For any questions regarding VSClinical’s cancer interpretation capabilities, please contact us at [email protected].
References
- Kundra, Ritika, et al. “OncoTree: a cancer classification system for precision oncology.” JCO Clinical Cancer Informatics 5 (2021): 221-230.
- Sioutos, Nicholas, et al. “NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information.” Journal of biomedical informatics 40.1 (2007): 30-43.