We have been building up to the final release of VarSeq 2.6.0 for a couple of weeks now, but we are excited to announce that 2.6.0 is officially available! VarSeq 2.6.0 is an exciting release as this version of VarSeq features the introduction of VSPGx, offering a complete pharmacogenomic workflow, including data import, variant analysis, and report generation. We have already generated TONS of helpful resources for learning about VSPGx and how to build a workflow that suits your lab. Check out these amazing resources below!
- VSPGx Product Page
- Webcast Introducing VSPGx: Pharmacogenomics Testing in VarSeq
- Pharmacogenetics eBook
- Upcoming Webcast: VarSeq PGx: A User’s Perspective on April 17th!
Even more exciting is that there is more to VarSeq 2.6.0 than just a brand-new genomic data analysis tool. Allow me to discuss some other cool developments that 2.6.0 brings to the table.
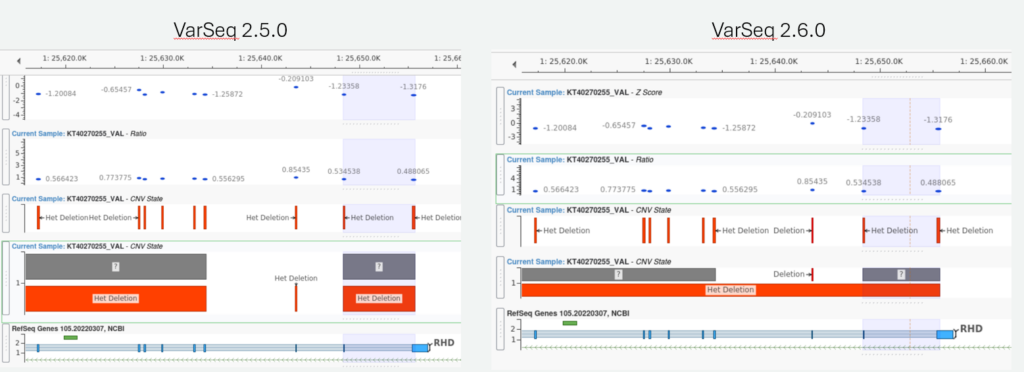
For all of our CNV users out there, VarSeq 2.6.0 comes with some serious upgrades and improvements to the CNV caller. Starting with the LOH caller, this algorithm has been optimized to read VAF and GQ values in each chromosome for all project samples at once, significantly improving the algorithm’s speed and efficiency. The CNV caller itself has been improved to better handle large CNV events, primarily by merging adjacent CNV calls when there are no targets between the CNV calls. Figure 1 shows an example of this new CNV event merging behavior. Other improvements were made to the CNV caller to handle calling CNV events in the X and Y chromosomes.
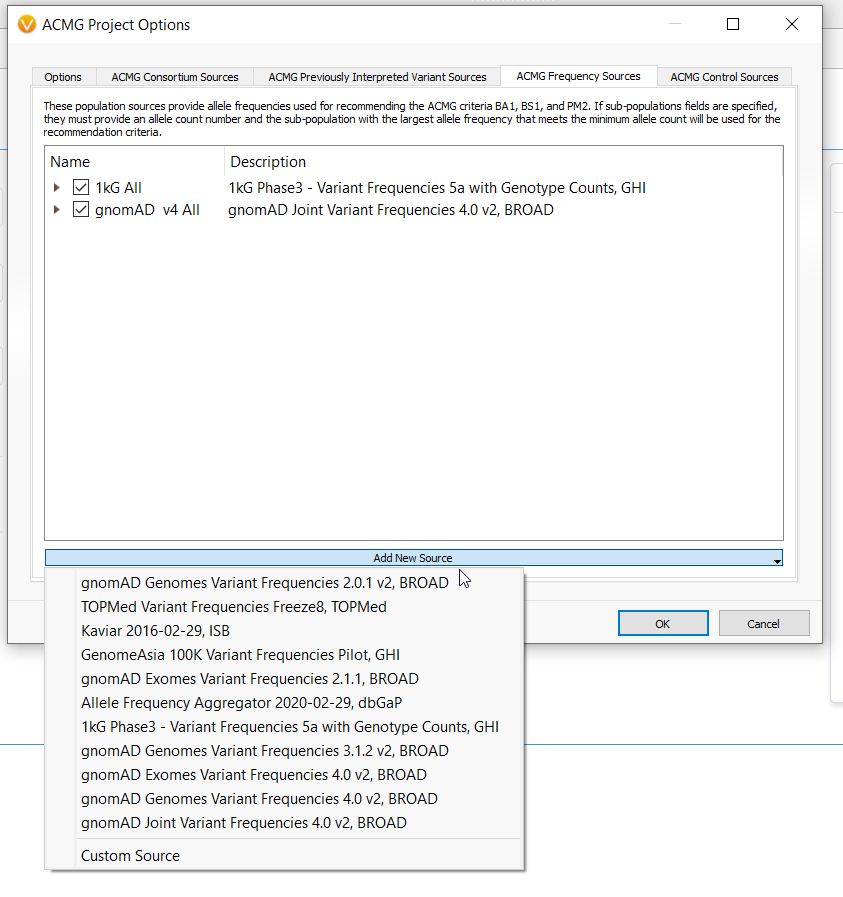
A few months ago, we curated the gnomAD v4 annotation sources. These annotations could be used in VarSeq 2.5.0 to annotate variants but to get these tracks integrated with the ACMG/Cancer Classifier algorithms and VSClinical came with some manual work. VarSeq 2.6.0 has incorporated the gnomAD v4 “Joint” tracks as default dependencies for the Classifier algorithms and has the auto-mappings for all gnomAD v4 tracks so they can easily be integrated with VSClinical (Figure 2). The “Joint”
tracks have also been added to all of our shipped project templates to get everyone started off on the right foot with population frequency analysis.
Several other fixes and features have made their way into VarSeq 2.6.0, so check out the release notes to get the full scoop on the release! Our amazing support team ([email protected]) is available to help you upgrade to the latest release, help set up your workflows, or answer any questions you might have. Cheers to VarSeq 2.6.0!