Sentieon — High-Performance Alignment & Variant Calling
Sentieon provides high-computing efficiency, fast turnaround time, and 100% consistency for secondary analysis NGS. Sentieon DNAscope delivers improved accuracy through enhanced active region detection, powerful local assembly of reads, and pre-trained machine learning models for both short-read and long-read sequencers — bundled directly with Golden Helix tertiary analysis.
Secondary analysis NGS is the critical computational phase that transforms raw sequencer output into structured variant data. It receives millions of short DNA reads in FASTQ format and produces a catalog of genetic differences relative to a reference genome, typically in VCF format.
Achieving high-throughput in clinical genomics requires variant calling software that balances speed, accuracy, and reproducibility. Legacy pipelines often face bottlenecks in processing whole-genome and whole-exome datasets, leading to extended turnaround times and hardware scaling challenges.
Golden Helix integrates Sentieon as a high-performance DNA sequencing pipeline engine, providing alignment, deduplication, and variant calling in a single optimized solution. By implementing GATK and MuTect2 mathematical models in low-level C and assembly language, Sentieon delivers 10X-50X performance gains while maintaining mathematical equivalence. Sentieon DNAscope goes further with pre-trained machine learning models that improve accuracy through enhanced active region detection and more powerful local assembly of reads for both short-read and long-read sequencers.
What Is High-Performance Variant Calling?
Modern genomic research demands a high-performance variant calling approach that can scale to national-level genome projects. Sentieon achieves this through optimized software implementation of the GATK Best Practices workflows.
- BWA Alignment: Mapping sequencing reads to the reference genome using BWA-MEM or BWA-MEM2 algorithms with enhanced computational efficiency.
- Deduplication: Identifying PCR artifacts and marking redundant copies to improve variant calling accuracy and reduce data volume.
- Variant Calling: Statistical models examine read pileups to determine genotypes using Bayesian inference for SNVs and Indels. DNAscope incorporates pre-trained machine learning models for improved accuracy.
Why Sentieon Outperforms Legacy Tools
Sentieon provides the most efficient secondary analysis solution for high-throughput environments where processing speed and consistency are critical.
- Mathematical Equivalence: Implements the exact same mathematics as BWA-GATK and MuTect2, achieving mathematically equivalent results with massive throughput gains.
- DNAscope ML Caller: Pre-trained machine learning models provide improved accuracy through enhanced active region detection and more powerful local assembly of reads for both short-read and long-read sequencers.
- 100% Deterministic: Eliminate run-to-run variation. The same input always produces the exact same output, which is critical for clinical validation.
- Generic CPU Support: Pure software solution that runs on standard CPUs—no specialized GPUs or expensive hardware accelerators required.
What to Look for in Secondary Analysis Software
Speed & Efficiency
The software should maximize CPU utilization to deliver 10X-50X improvements in core-hours over standard Java-based pipelines.
Reproducibility
Deterministic results with no downsampling in high-coverage regions ensure consistent variant calls for clinical delivery.
Scalability
Support for joint calling on 100,000+ samples without intermediate file merging, enabling massive cohort studies.
Somatic & Germline
Unified support for DNAseq (germline) and TNseq (tumor-normal somatic) variant calling in one engine.
Tertiary Synergy
The secondary analysis should integrate directly with tertiary analysis for a seamless "FASTQ to Report" workflow.
Deployment Flexibility
Deploy anywhere: on-premises workstations, centralized servers, or air-gapped secure networks.
The Complete Pipeline: Sentieon + VarSeq
Golden Helix provides a complete bioinformatics pipeline designed to receive data directly from the sequencer and carry it through to clinical reporting. Sentieon processes the raw FASTQ data into a VCF, which is then imported into VarSeq for filtering and annotation.
Unmatched Speed
Achieve 10X faster FASTQ-to-VCF and up to 50X faster BAM-to-VCF processing compared to standard BWA-GATK pipelines.
Clinical Grade Accuracy
Sentieon implements mathematically equivalent GATK models for proven accuracy. DNAscope goes further with ML-optimized calling that improves sensitivity and specificity for both short-read and long-read data.
Seamless Automation
Integrate Sentieon into VSPipeline for full automation from raw sequencing reads to clinician-ready PDF reports.
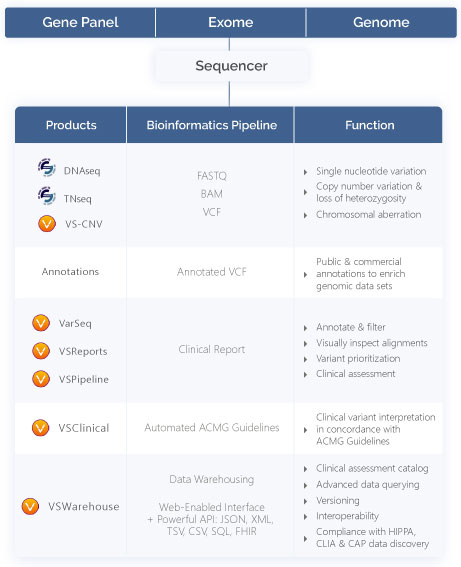
"Sentieon provides the upstream secondary analysis that feeds directly into VarSeq's tertiary interpretation engine."
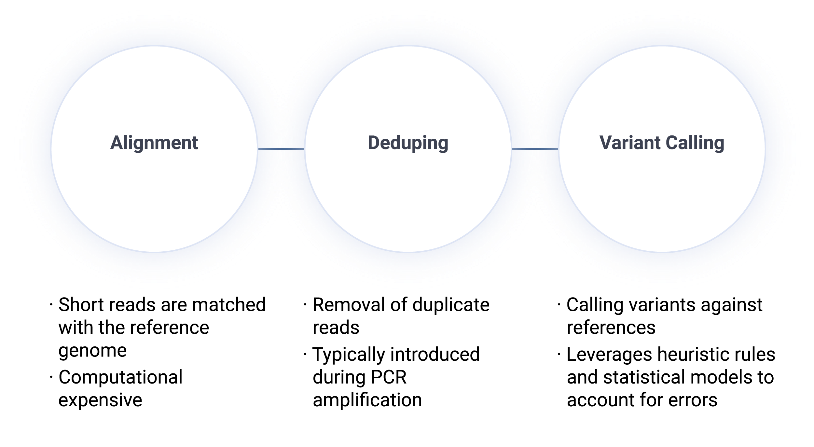
The Sentieon Secondary Analysis Workflow
Read Alignment
Map sequencing reads to the reference genome using highly efficient BWA-MEM or BWA-MEM2 algorithms.
Deduplication
Identify and mark PCR artifacts to ensure variant calls are based on unique biological fragments.
Variant Calling
Apply Bayesian models (DNAseq) or ML-optimized calling (DNAscope) to identify SNVs and Indels with deterministic, 100% consistent results.
VCF Generation
Produce standard VCF files ready for clinical-grade tertiary analysis and interpretation in VarSeq.
Enterprise Scaling for Population Genomics
Sentieon is designed for massive datasets, supporting joint calling on 100,000+ samples without intermediate file merging.
- Pure Software Solution: Runs on standard CPUs—no specialized GPUs needed.
- No Downsampling: Uses every read even in ultra-high coverage regions.
- WGS & WES Ready: Optimized for whole-genome and whole-exome scale data.
Germline & Somatic Solutions
Germline DNAseq & DNAscope
Complete solution for germline SNV and Indel detection. DNAseq provides mathematically equivalent GATK results at 10X speed, while DNAscope uses ML models for even higher accuracy on both short and long reads.
Variant InterpretationSomatic TNseq
Tumor-normal pair somatic variant detection matching MuTect and MuTect2 mathematics with significantly improved efficiency.
Somatic SolutionsPopulation Joint Calling
Scale to national genome projects with joint calling for 100,000+ samples. Deterministic results ensure data consistency across cohorts.
Whole Genome AnalysisFrequently Asked Questions
Are Sentieon results compatible with GATK?
Yes. Sentieon's DNAseq implements the exact same mathematical models as Broad Institute's Best Practice Workflows, achieving mathematically equivalent results with massive throughput gains. DNAscope goes beyond GATK equivalence by incorporating pre-trained machine learning models for improved accuracy on both short-read and long-read data.
Does Sentieon require GPUs for acceleration?
No. Sentieon is a pure software solution optimized for standard CPU architectures. It delivers 10X-50X performance gains without requiring specialized hardware like GPUs or FPGAs.
What is meant by "100% deterministic results"?
Unlike some bioinformatics tools that may produce slightly different results across multiple runs due to multi-threading randomness, Sentieon ensures that the exact same input always produces the exact same output.
Can I automate Sentieon within my existing pipeline?
Yes. Sentieon is highly modular and can be integrated into VSPipeline or custom automation scripts, enabling a hands-off workflow from FASTQ generation to clinical reporting.
Secondary Analysis Insights & Webcasts
Learn how to optimize your NGS pipeline for speed and accuracy with Sentieon and VarSeq.
Ready to Accelerate Your Secondary Analysis?
Join high-throughput clinical labs and research centers worldwide using Sentieon for high-performance variant calling.