
As whole-genome sequencing becomes routine in clinical practice, variant interpretation remains both the opportunity and the obstacle. VarSeq users can efficiently process and report individual samples using extensive annotations and automated classification tools—but even the most comprehensive databases can leave gaps. Some variants simply aren’t found in ClinVar or ClinGen, leaving analysts without clear direction. This is where Genomenon’s Mastermind and CKB integration changes the landscape, delivering curated literature evidence directly within VarSeq to support confident, evidence-based interpretation.
This blog highlights a real-world example in which, without access to the Mastermind database, a variant’s classification would have remained at uncertain significance—illustrating the practical value and clinical impact of our strategic partnership with Genomenon.
Deep Intronic Variant in PAH
One major benefit to whole genome analysis is the capture of deep intronic variants. In many cases, you may rely on splice prediction tools to determine if the variant is either impacting a canonical splice site or potential introducing a novel site. Figure 1 illustrates this position of this particular PAH variant that is approximately 250 bases from the nearest splice motif.
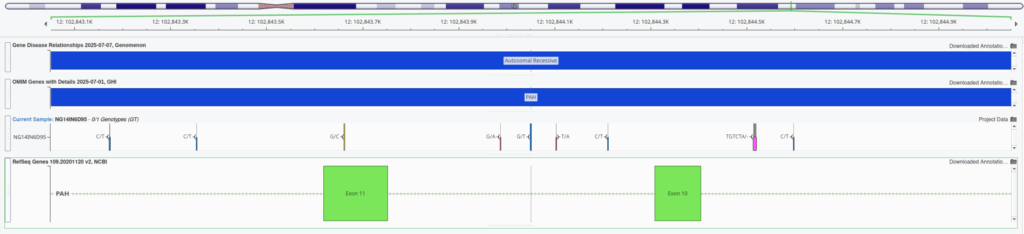
The issue is seen in Figure 2 when annotating this variant, it is not seen in ClinGen, nor ClinVar and it is likely too deep to detect as a novel splice variant even with sophisticated tools such as SpliceAI. The resulting ACMG classification is that the variant is not only left with sparse evidence to assign criteria such as PM2 for its rarity, but is even predicted to not impact splicing with BP7.

However, when integrating the Genomenon Mastermind database, in Figure 3, the autoclassification is upgraded to Pathogenic due to the curation of relevant literature (PMID 36849017). The result of factoring for the functional study not only allows for PS4 but most critically PVS1 where the variant results a deleterious psuedo-exon impact on protein function.

Conclusion
The case of this deep intronic variant underscores the critical role curated databases like Genomenon’s Mastermind play in clinical genomics. Falling outside the reach of common clinical databases and splice prediction tools, variants like these highlight the importance of using every available resource in the pursuit of supporting evidence. Thanks to Genomenon’s continued efforts in curating variant data and associated literature, these databases ensure that no clinically relevant variant goes unnoticed. Seamlessly integrated into Golden Helix’s ACMG classifier and VSClinical interpretation software, Mastermind enables users to streamline variant interpretation and reporting—turning complex discoveries into confident, evidence-backed conclusions.