It doesn’t take much effort to find articles discussing the value of Next-Generation Sequencing (NGS). There is a consistent tone amongst authors that implementing NGS pipelines are critical for clinical efficiency in both hereditary disorders and somatic. However, NGS strategies do not come without their own challenges. Challenges include not only the detection and calling of high quality/probability variants from a secondary analysis perspective, but also huge hurdles in the tertiary realm processing the data and reaching conclusions. Tertiary obstacles include handling scale of data, i.e., gene panels with hundreds of variants versus whole genome with millions, efficient filtration to clinically relevant variants, and even the thorough review of all relevant variant evidence following robust guidelines. Now let us assume we successfully navigate through these stages. We develop a rock-solid secondary pipeline generating our variants, which then pass through a comprehensive and relevant filtering logic isolating a single pathogenic variant relevant for the patient’s disorder and reach a final classification/interpretation/clinical report. There is still another challenge to consider post evaluation. How do we capture all this hard work and final interpretation? Where do we record this variant for future review in case classifications change? How can I leverage the known variants processed in the past to add context for future variants in the same gene?
Luckily, Golden Helix provides solutions for every challenge mentioned here and more. The purpose of this blog series will be to demonstrate our capabilities overcoming the last challenges mentioned, the storage of the final variants, and how to optimize your workflows with this growing knowledge base. We will be first discussing this in the context of GHI assessment catalogs, storing SNV, and indels processed through VSClinical’s automated ACMG guidelines. Part II of this blog will cover the same considerations but in the context of Copy Number Variants. Lastly, Part III will showcase the simplest infrastructure by storing these catalogs, variants, projects, reports, and other relevant genomic data in VSWarehouse. Let us begin by highlighting the components of variant classification following ACMG guidelines in VSClinical.
We briefly covered the elements of tertiary analysis, which is completed with VarSeq. Within VarSeq, users can import all variant data contained in the VCF. VarSeq then provides multiple algorithms and annotations supplying relevant fields used to help filter variants down to those that are clinically relevant. The next step is to then review the variant following the automated ACMG and AMP guidelines in VSClinical (Figure 1). The VSClinical terminal is comprised of multiple tabs that compartmentalize relatable criteria. The population tab consists of the criteria related to variant allele frequency among germline population catalog (PM2). In comparison, the Gene Impact tab contains criteria relevant to the measured influence of the variants impact on the gene (PVS1 for loss of function variants). The Studies tab then contains the criteria selection based on previous clinical submissions interpretations available through multiple literature sources (PP5 for variants previously classified pathogenic in ClinVar).
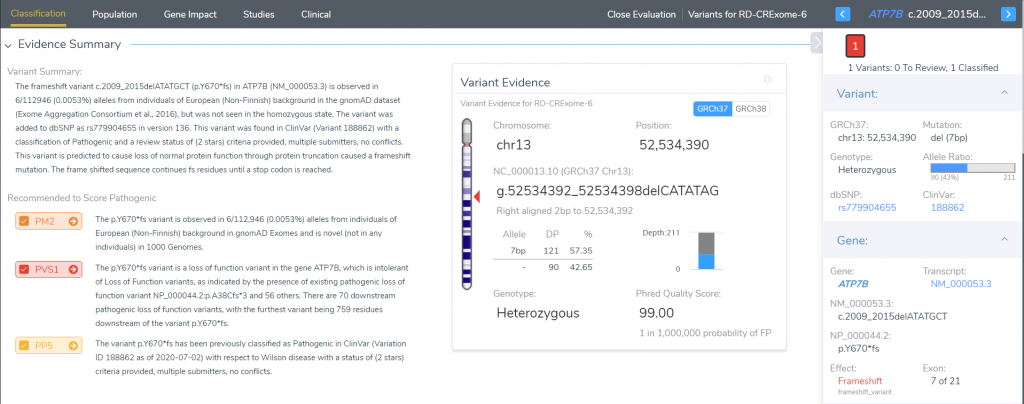
Figure 1. Screenshot of the VSClinical terminal for the ACMG guideline processing of variant evidence to reach final classification
Once a variant has been evaluated, the classification and interpretation is the next step. Figure 2 is a snapshot of the interpretation build for the variant in Figure 1. However, you’ll notice the Accept Previous icon at the bottom. Essentially, this variant has been reviewed before in another sample, and that previous classification can be leveraged for future samples. Not only does storing these classifications save time for future variant evaluations, but it also is easy to be reviewed for accuracy and can be referenced for changing classifications in the future.
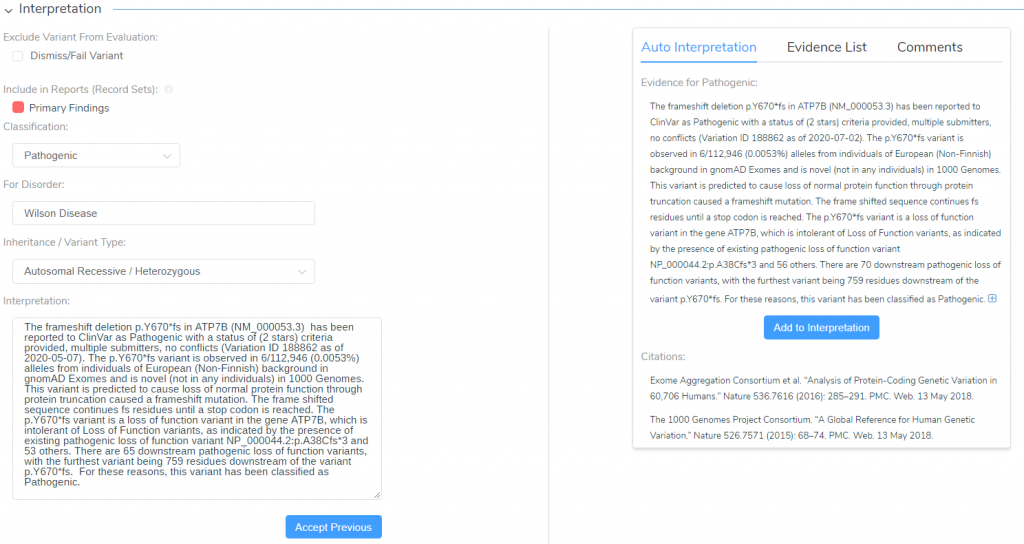
Figure 2. Final classification/interpretation section of VSClinical, showing a previously captured classification for the current variant
Every VSClinical user will create a catalog to store these classifications and interpretations. Figure 3 shows the window users will access when setting up their catalog. Once made, the catalog will capture a number of fields the user can leverage from sample to sample. Figure 4 shows the fields automatically built and filled across all evaluated variants.

Figure 3. Setting up and selecting the preferred assessment catalog for variant classification capture

Figure 4. List of all fields auto-populated with variant and sample data following ACMG evaluation
As you can see, there are many fields to work with; all exportable, all possible to include in the filter chain. However, regarding tracking evolving classifications, not only does the catalog store the previous classification, but it also captures the author, interpretation, sample name, disorder, and last edited details. This helps reference alternate interpretations but also those that change with new evidence from the annotations. We will explore more options for leveraging changing classifications when looking at this from the VSWarehouse perspective in part 3 of this blog series. The purpose of this blog has been more conceptually based so to see these tools in action; please reach out to [email protected] for a one-on-one training should you have any further questions, or enter them below in the comments section!