In this blog post, I’ll highlight the benefits of using VSPipeline to automate tertiary analysis in VSWarehouse. Users can now run VSPipeline project creation as a standalone task or a final step in a bioinformatic workflow without writing code. Simply use one of our shipped VarSeq projects or create your own, then automate that project as a template that can… Read more »
…so we added some automation to your automation so you can automate while you automate! Automation has been a hot topic recently and for all the right reasons. As we (proudly) watch our customers increase their sample and data volume, we are constantly seeking to provide tools to reduce click rate and optimize throughput. Furthermore, with all of the new… Read more »
As a lab or group scales the number of NGS samples analyzed, it is important to automate the sample analysis pipeline from the sequencer to the point where it is ready for a variant scientist or lab personnel to follow the interpretation workflow and draft a clinical report. VSPipeline leverages the core VarSeq capability to create reproducible test-specific workflows through… Read more »
VSPipeline is becoming a very popular tool among VarSeq users as it is essential for creating repeatable clinical workflows that can be executed in automated fashion. Since VSPipeline is a command-line tool, I think it would be helpful to discuss some of the best practices along with helpful tips for getting the most out of VSPipeline. Some of you may be less familiar with VSPipeline, so I want to cover how to set up the first run along with sharing the helpful tips as they arise. … Read more »
VSPipeline is a command-line interface that will provide high throughput environments the ability to tap the full power of VarSeq’s algorithms and flexible project template system from any command-line context, including the existing bioinformatics pipeline. This feature is a great resource for analyzing large sample volumes as it automates importing and annotating your data, which can help streamline your analysis… Read more »
The power of VSPipeline is in it’s ability to automate VarSeq workflows. Using VarSeq to create a pipeline template is great because it allows you to dial in the applied filters as well as interactively organize the annotations and applied algorithms. Automating a workflow with VSPipeline is straightforward when beginning with an existing project. However, there are several steps that… Read more »
Today, we are proud to announce our collaboration with Fulgent Genetics, a CLIA certified molecular diagnostics lab. Fulgent offers more than 4,000 single gene tests among others and will implement VSPipeline to help speed up their analysis and interpretation process. On our quest to enable precision medicine, we look forward to working with Fulgent and other diagnostics labs in the… Read more »
Recently, Golden Helix, Inc. announced the addition of VSPipeline to our VarSeq software suite. VSPipeline is a command-line interface that will allow high throughput environments the ability to tap the full power of VarSeq’s algorithms and flexible project template system from any command line context, including existing bioinformatics pipeline. So, what is the big deal? Here are the top five… Read more »
When it comes to cervical cancer, early detection and timely analysis can literally be life-saving. Cervical cancer is the fourth most common cancer in women worldwide, with an estimated 660,000 new cases and around 350,000 deaths each year. Early detection through screening and quick diagnostic support drastically improves outcomes because, when caught early, cervical cancer is among the most preventable… Read more »
When quality metrics generated during secondary analysis are not incorporated into interpretation and reporting, variants are reported without the important context that could affect confidence in the call. The Golden Helix software suite has been designed to help you bring in quality metrics from secondary analysis through to interpretation and reporting. When using VarSeq, it’s essential to leverage the software’s… Read more »
Short-read sequencing often fails to capture clinically actionable information from challenging genomic regions. While long-read sequencing now enables accurate and comprehensive detection of complex variants, there are still regions of the genome that remain notoriously difficult to analyze. Unfortunately, many of these regions contain genes that are highly relevant to precision medicine. Consider CYP2D6 and CYP2C19, which guide pharmacogenomic dosing… Read more »
At Golden Helix, we’re proud to see how researchers worldwide continue using our software to accelerate discovery and improve clinical decision-making. These October 2025 publications highlight how VarSeq is advancing genetic research across cancer, hereditary disease, and molecular diagnostics. From identifying new variant mechanisms to expanding our understanding of genotype–phenotype relationships, these studies demonstrate how Golden Helix tools empower teams… Read more »
Recently, Golden Helix attended the ASHG 2025 conference, where we had the opportunity to showcase our partnership with Genomenon. This collaboration took center stage at both our booth and Genomenon’s CoLab session, Automating Genomic Workflows: Cutting Interpretation Time, Accelerating Turnaround, and Increasing Diagnostic Yield. Our discussions centered on a problem that every clinical lab faces as variant volumes continue to… Read more »
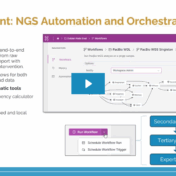
As Next-Generation Sequencing (NGS) becomes central to modern clinical diagnostics, laboratories are managing more data than ever before, and turning that data into actionable insight requires both precision and scalability. In our recent webcast, Next-Gen Sequencing at Scale: Transforming Per-Sample Outcomes into Institutional Knowledge with VSWarehouse, we explored how Golden Helix’s software ecosystem streamlines NGS analysis, bridging the gap between… Read more »
Short Tandem Repeats (STRs) are increasingly recognized as clinically important genetic variants, contributing to a wide range of rare inherited disorders. Historically, the analysis and clinical reporting of STRs have been challenging due to limitations in both sequencing technologies and interpretation frameworks. With the advent of long-read sequencing technologies and the advanced secondary analysis pipelines provided by PacBio, Oxford Nanopore,… Read more »
At Golden Helix, we get to see firsthand how our software enables groundbreaking research around the world. Our tools are central to real-world discoveries, from cancer studies and prenatal diagnostics to uncovering new genetic links in rare diseases. These publications from August 2025 showcase how VarSeq and VSPipeline are helping researchers push the boundaries of what’s possible—automating workflows, streamlining variant… Read more »
As a new member of Golden Helix’s Field Application Services team, I’ve recently been diving into the capabilities of VarSeq and our broader software suite – an experience every new VarSeq user goes through. If I could schedule a training call with my former self, there are some key points I’d share to help hit the ground running. Here are… Read more »
Managing long‑read secondary analysis used to mean wrestling with Cromwell servers, custom compute nodes, and a tangle of WDL files. VSWarehouse 3 removes that burden. Drop your PacBio output into a watched folder, and our platform does the rest: spinning up compute, tracking every task, and handing you fully annotated results ready for clinical interpretation. Built‑In Workflows, Ready to Launch To… Read more »
VarSeq, when used with VSPipeline, automates carrier screening analysis, allowing labs to run them in a parallelized, economical fashion.
Thank you to everyone who joined our recent webcast, “Powering Genomic Workflows with Upgraded Catalogs in VSWarehouse and VarSeq 3,” presented by Gabe Rudy on April 23rd, 2025. We appreciate the engagement and interest in the latest advancements to our platform. For those who missed it or need a recap, the session focused on the pivotal role Catalogs play as… Read more »