Genetic Data Warehousing e-Book In our webcast last week, we announced the upcoming release of VSWarehouse, a scalable genetic data warehouse for VarSeq. Genetic data warehousing becomes more important as Next-Generation Sequencing is taking off in the clinic, creating significant data management issues for clinicians, scientists and IT professionals alike. How can we retain the massive amounts of data coming out… Read more »
Compound Heterozygous Workflows: Including a 2nd Affected Child Looking for Compound Heterozygous regions for a trio is fairly straight forward in VarSeq, we include this workflow in our shipped Exome Trio Template. An example of which is included with our Example Projects which can be found by going to File > Example Projects > Example YRI Exome Trio Analysis. But… Read more »
So, why are we launching a new data warehouse product? Why did we build VSWarehouse? According to Grand View Market Research, the next generation sequencing (NGS) market size was $2.0 billion (USD) globally in 2014. This number is expected to grow from 2015 to 2022 at an annual rate of about 40%. What drives this phenomenon is the increasing number… Read more »
Golden Helix in 2016 We had a terrific year 2015. It was the year in which we got serious about the clinical testing market. We successfully continued on the path of attracting more referenceable clients such as University of Iowa, Baby Genes, Prevention Genetics and many more. We rounded out our VarSeq suite by adding more clinically relevant features and… Read more »
The most common use of the VarSeq Match Gene List algorithm of course is to determine if the variants in your data set are contained within your genes of interest. As an example of this, say you are working with a whole exome trio and only want to consider those variants that are contained within the 56 genes recommended by… Read more »
Probably one our most popular public annotation sources we curate and update is the database of Non-Synonymous Functional Predictions (dbNSFP). In it’s recent update, it has expanded the predictions to include FATHMM-MKL and VarSeq now incorporates this new prediction into its voting algorithm of now 6 different discrete predictions per variant. You can update to dbNSFP 3.0 using the built-in… Read more »
As VarSeq’s adoption has grown among analysts using whole exome data to diagnose rare diseases, a couple of family designs outside of the common trio of an affected child and both parents have come up frequently. While having both parents provides the maximum power to discover de novo mutations and recessively inherited variants, it is not always possible to contact… Read more »
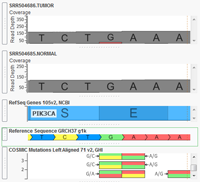
With the release of VarSeq 1.3.1 we have included a new demo project to showcase a single tumor-normal pair analysis workflow. The project can be accessed through VarSeq and VarSeq Viewer by going to File > Open Example Projects > Example Tumor-Normal Pair Analysis. This project contains an exome pair (Normal-N990005 and Tumor-T990005) from the Gastric Cancer study Exome sequencing of… Read more »
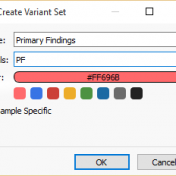
With the release of VSReports, we added the ability to “select” rows of your filtered output (often variants, but potentially things like coverage regions or genes) with a new feature dubbed “Record Sets”, but more often described as “colored checkboxes” for your tables. Although necessary for the important task of marking primary, secondary or other sets of variants for a… Read more »
As VarSeq gains in popularity, we want to give Viewers and customers alike the opportunity to look at projects that are completed from start to finish. To this end, VarSeq (and VarSeq Viewer!) currently comes with two demonstration projects, Example TruSight Cardio Gene Panel and Example YRI Exome Trio Analysis. To access these projects from the VarSeq start page go to… Read more »
Our webcast yesterday featured two clinical workflows and and the ease in moving from an unfiltered variant file to a clinical report containing the variants of interest using VarSeq and VSReports. There were several great questions and I wanted to pass on a few of particular interest. Question: Are annotation sources included in VarSeq for free?
While VarSeq comes with a number of starter workflows that are stored as templates, customers also have the option of creating filter chains from scratch; analyzing a single exome may require you to do exactly that. In this blog, I’ll go through analyzing a single exome and generating a list of variants for further study. After importing the variant data… Read more »
The 65th annual ASHG in Baltimore will be another exciting one. We at Golden Helix have been very busy this year making great improvements to both SVS and the VarSeq software and we look forward to showcasing them during our in-booth demos. In particular, we will launch two new additions to the VarSeq software suite; VSReports and VSWarehouse. VSReports brings highly customizable clinical… Read more »
Yesterday, our VP of Product and Engineering, Gabe Rudy, presented VSReports to the Golden Helix community for the first time in a live webcast; Authoring Clinical Reports in VarSeq. It was an excellent presentation. Gabe highlighted VSReports’ ability to take the output of tertiary analysis to a customized clinical grade report in one click. He also gave an overview of… Read more »
The next release of VarSeq will ship a new product that is highly relevant to our customers in clinical testing labs. Via VSReports, VarSeq now has the ability to generate clinical-grade reports. These reports are fully customizable, containing focused and actionable data. VS Reports ships with report templates that are modeled off of the ACMG guidelines, the de-facto gold standard… Read more »
When it comes to down to it, the genomic variants we collect in a research and clinical setting are impossible to interpret without that important link of how genes are related to phenotypes. Indisputably, the Johns Hopkins project to catalog all evidence related to inheritable Mendelian diseases is our best repository of this evidence. Online Mendelian Inheritance in Man (OMIM)… Read more »
I was definitely an early adopter when it comes to personal genomics. In a recent email to their customer base announcing their one millionth customer, they revealed that I was customer #44,299. And I have been consistently impressed with the product 23andMe provides through their web interface to make your hundreds of thousands of genotyped SNPs accessible and useful. It… Read more »
Today, we are proud to announce our collaboration with Fulgent Diagnostics, a CLIA certified molecular diagnostics lab. Fulgent offers more than 4,000 single gene tests among others and will implement VSPipeline to help speed up their analysis and interpretation process. On our quest to enable precision medicine, we look forward to working with Fulgent and other diagnostics labs in the… Read more »
VarSeq now supports analysis of paired Tumor/Normal samples! Tumor/Normal support has been one of the most common feature requests for VarSeq since it was launched late last year, and we are excited to make this functionality available to all of our VarSeq users in the latest update (version 1.1.4). VarSeq is a powerful platform for annotation and filtering of DNA… Read more »
Recently, Golden Helix, Inc. announced the addition of VSPipeline to our VarSeq software suite. VSPipeline is a command-line interface that will allow high throughput environments the ability to tap the full power of VarSeq’s algorithms and flexible project template system from any command line context, including existing bioinformatics pipeline. So, what is the big deal? Here are the top five… Read more »