Golden Helix Featured in 20 Most Promising Biotech Technology Solution Providers for 2016! This month, CIO Review released a Biotech Technology Special in which Golden Helix was named among the 20 most promising biotech technology solution providers for 2016. Read the interview with our CEO, Andreas Scherer here: Empowering Precision Medicine through Next-Gen Technology Our customers are monumental in this… Read more »
Wednesday, April 6th 12:00 pm EDT As the number of samples and associated data volume in a testing lab increases, it becomes imperative for labs to leverage state of the art warehousing technology that not only organizes data, but also aides and enables researchers and clinicians to perform further analysis, and ongoing research. Built on the algorithms and high-performance storage… Read more »
Concepts and Relevance of Genome-Wide Association Studies Genome-Wide Association Studies continue to be a very effective method for determining the underlying cause of disease, Golden Helix is happy to share our long-standing knowledge with the community. We are very happy to announce that “Concepts and Relevance of Genome-Wide Association Studies”, a paper surrounding GWAS was recently published in Science Progress… Read more »
We, at Golden Helix, would like to take the time to highlight one of our long time customers on her recent success. Since the beginning of 2015, Dr. Kazima Bulayeva of the Vavilov Institute of General Genetics (VIGG), has published in 10 scientific publications including two times Nature Genetics. We are very excited to share the research she is performing using the… Read more »
Headed to Tampa! For the first time, Golden Helix will be attending the Annual Clinical Genetics (ACMG) meeting. This year, the meeting is being held in Tampa, Florida and we are excited to visit with our customers and meet new people in the field! Where to Find Us You can find the Golden Helix team in booth #435 in the… Read more »
Thank you to all who submitted an abstract in the 2016 Abstract Challenge, it was a great success! With more than 30 submissions and a wide variety of topics, selecting only 3 winners very difficult! With that being said, here are the top 3 winners for this year’s Abstract Challenge: Our 1st place winner is Dr. Folefac Aminkeng, Postdoctoral Fellow… Read more »
Scaling is in our DNA: Making Genomics Accessible One of the things I absolutely love about the work we do at Golden Helix is keeping up with the changes in data analysis driven by the iterative and generational leaps in technology. But one thing has always been a constant since day one: we break preconceived notions of what scale of… Read more »
New SVS Meta-Analysis Example Project The latest version of the GWAS E-book featured a chapter on Meta-Analysis. We are pleased to make the project used in this chapter available as an example project for SVS (SNP & Variation Suite). This blog post will walk you through the analysis steps used in the example project. This information is also contained in… Read more »
Compound Heterozygous Workflows: Including a 2nd Affected Child Looking for Compound Heterozygous regions for a trio is fairly straight forward in VarSeq, we include this workflow in our shipped Exome Trio Template. An example of which is included with our Example Projects which can be found by going to File > Example Projects > Example YRI Exome Trio Analysis. But… Read more »
With the release of our updated GWAS E-book, we have recently updated the GWAS example project (SNP Genome-Wide Association Tutorial – Complete). This updated project includes more details about how spreadsheets were generated, how to generate plots and which images were used for the GWAS E-book. This information can be found in the User Notes view in the project navigator and… Read more »
Golden Helix was named one of the Top 10 Analytics Solutions Providers for 2016 by Pharma Tech Outlook! We are very excited to be honored in this month’s issue of Pharma Tech Outlook as one of 2016’s top ten Analytics Solutions Providers. Read the interview with our CEO, Andreas Scherer here: Empowering Biologists to Perform Complex Analyis We want to… Read more »
Recent Customer Publications We hope everyone had a Merry Christmas! The Golden Helix team is looking forward to the start of a new year, but first we wanted to share our last round of customer publications from 2015. Here’s to many more in 2016! Bishwa Sapkota and Dharambir Sanghera of the Oklahoma University Health Sciences Center and colleagues published The Genome-wide Association Study of… Read more »
The 65th annual ASHG in Baltimore will be another exciting one. We at Golden Helix have been very busy this year making great improvements to both SVS and the VarSeq software and we look forward to showcasing them during our in-booth demos. In particular, we will launch two new additions to the VarSeq software suite; VSReports and VSWarehouse. VSReports brings highly customizable clinical… Read more »
Yesterday, our VP of Product and Engineering, Gabe Rudy, presented VSReports to the Golden Helix community for the first time in a live webcast; Authoring Clinical Reports in VarSeq. It was an excellent presentation. Gabe highlighted VSReports’ ability to take the output of tertiary analysis to a customized clinical grade report in one click. He also gave an overview of… Read more »
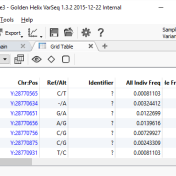
The support team at Golden Helix is always on-hand to help with your SVS and VarSeq needs. We get some questions more often than others, and this blog will answer some of the most common questions we’ve been seeing lately regarding VarSeq. A common question we receive is if data can be filtered from a locally kept set of variants… Read more »
Today we wanted to share a recent client case study to demonstrate how VarSeq has been implemented in a CLIA certified clinical laboratory. Please feel free to contact us if you have questions or if you would like to discuss VarSeq further at [email protected]. It is standard practice for newborns to be screened for genetic diseases before leaving the hospital to… Read more »
Today at Golden Helix, we are proud to announce our collaboration with MedGenome through an integration of OncoMD into our VarSeq software. Now VarSeq’s streamlined process of annotating and filtering variants will offer an added dimension. OncoMD is a comprehensive knowledge base of cancer-specific genetic alterations, and by incorporating it into VarSeq, users can access to 2 million plus annotated… Read more »
Over 650 GenomeBrowse licenses have been registered and downloaded since the beginning of 2015, and with so many people enjoying the utility of this freeware program, I wanted to showcase some advanced tips and tricks so you can get more out of GenomeBrowse! Under the Controls panel, when you’re clicked inside a data plot, there is a “Filter” tab. This… Read more »
This week, Dr. Jeffery Moore presented a webcast on the Molecular Sciences Made Personal. The webcast delved into Dr. Moore’s attempts to transform how they teach chemistry at the University of Illinois and demonstrated how he uses VarSeq with his students to examine exome data. The following are the questions asked by the attendees. Please feel free to reach out… Read more »
To say the announcement of Dan MacArthur’s group’s release of the Exome Aggregation Consortium (ExAC) data was highly anticipated at ASHG 2014 would be an understatement. Basically, there were two types of talks at ASHG. Those that proceeded the official ExAC release talk and referred to it, and those that followed the talk and referred to it. Why is this… Read more »