In the new Genotype Imputation tool that is coming soon to SVS, allele encoding is an important part of matching data between the target and the reference panels. If the same platform provider is being used, then A/B encoding can be used. However, it’s better to use the Reference/Alternate allele encoding associated with AGCT format to ensure accuracy. If an… Read more »
Since we released our Phenotype Gene Ranking algorithm in VarSeq, it has become a staple of the way people conduct their analysis. It allows for a combination of filtering with ranking to prioritize follow-up interpretations of analysis results. Our PhoRank algorithm will be available in our upcoming SVS release to also aid in the numerous research workflows performed on SNPs… Read more »
Genome-wide association study (GWAS) technology has been a primary method for identifying the genes responsible for diseases and other traits for the past ten years. GWAS continues to be highly relevant as a scientific method. Over 2000 human GWAS reports now appear in scientific journals. In fact, we see its adoption increasing beyond the human-centric research into the world of… Read more »
Join our upcoming webcast – Clinical Reporting Made Easy Wednesday, February 15th 12:00 pm EST Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the… Read more »
Just as we expected, 2017 has kicked off with a flurry of new publications by our customers. We even had a publication from a client using our VarSeq software! Congratulations to all, please take a look at some of the articles we have highlighted below: Reza Sailani of Stanford University and colleagues published Association of AHSG with alopecia and mental retardation… Read more »
With a properly defined wet-lab and bioinformatics process, we are able to zero in on clinically relevant variants. How does a lab report on the outcome of their analysis? We find that most laboratories conduct their variant classification based on the guidelines formulated by the American College of Medical Genetics (ACMG) for inherited diseases. The ACMG guidelines for variant classification… Read more »
It may have been easy to miss in the drum-beat of monthly annotation updates we do here at Golden Helix, but there are a couple of things that are very special about the January update to the ClinVar database: We added new fields including HGVS names of variants and citations in PubMed for variants ClinVar nearly doubled in size by… Read more »
Acute lymphoblastic leukemia (ALL) is the most frequently diagnosed cancer in children and one of the leading causes of death due to disease in children. Dr. Daniel Sinnett, along with Pascal St-Onge and their colleagues at Sainte-Justine University Health center have been investigating the molecular determinants of the disease to improve detection, diagnosis and treatment. One particular area of study… Read more »
The year 2017 is starting fast and furious for us here at Golden Helix. We just announced a new imputation capability for our SVS product. At the same time, members of our team are on the way to PAG in San Diego to network with our clients in the Plant and Animal community. We have a terrific plan in place… Read more »
Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the results of tertiary analysis into a customizable clinical report. To include the VSReports functionality in… Read more »
This Saturday, Plant & Animal Genome (PAG) XXV will kick off in sunny San Diego! Here in Montana, we have had a brutally cold winter thus far. I, like many of you, am looking forward to San Diego’s sunny, warm temperatures! Since last year’s PAG, SVS has had a number of updates including the integration of BEAGLE for imputation (coming soon!),… Read more »
Golden Helix closed out 2016 with a great honor; in December, The Silicon Review Magazine released a special edition naming Golden Helix as one of the fastest-growing technology companies for 2016. You can read the interview with our CEO, Andreas Scherer, Ph.D, here: Delivering industry-leading analytic software and services. We continue to believe that our customers are paramount in this honor, as… Read more »
Our webcast series for 2017 is starting off by giving the Golden Helix community their first look at our addition of genotype imputation into SVS! On Wednesday, January 11th, Gabe Rudy will discuss how users can now run their genotype phasing and imputation on human and animal data as part of their SVS analytics workflow. Wednesday, January 11th @ 12:00 PM, EST Genotype imputation… Read more »
It sure is feeling like Christmas time in Montana with the piles of fluffy snow and negative temperatures! We are wrapping up the month with a few more publications from our clients, and we couldn’t be happier with how many articles were published in 2016! Congratulations to everyone who was able to get it done this year, and we are looking… Read more »
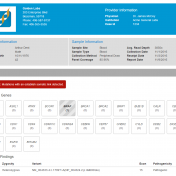
Variant interpretation is an integral part of any workflow that results in some decisions being made about the validity and suspected functional impact of a variant in a given sample and their presenting phenotypes. The VarSeq Assessment Catalog functionality is designed to assist the VarSeq user in streamlining this process. To include this functionality in your workflow, you will first… Read more »
It is time to kick off our fourth annual abstract challenge! Our mission at Golden Helix is to enable Precision Medicine. We accomplish this by creating powerful software that enables both researchers and clinicians to complete complex analysis. This competition allows us to enable discovery by awarding software to those making an impact in the field. The competition has been… Read more »
ExAC CNVs were released publicly with a recent publication, providing the full set of rare CNVs called on ~60K human exomes. While there are many public CNV databases out there, this is the first one that was derived from exome data, and thus includes both extremely rare and very small CNV events. With the recent release of Golden Helix’s CNV calling… Read more »
Dr. Laura Li and her colleagues at the Children’s Hospital Los Angeles (CHLA) are working to determine the underlying genetic causes of Optic Nerve Hypoplasia (ONH), which is still unclear. ONH is the absence or under-development of the optic nerve and is currently the leading ocular cause of vision impairments and blindness in young children. ONH can also be combined… Read more »
We hope our Golden Helix community had a delicious Thanksgiving, and several of our customers had an additional reason to celebrate because they published this month, including one book publication! The success is overwhelming and we are happy to share it with you each month: Randovan Kasarda of the Slovak University of Agriculture and colleagues published Genetic Divergence of Cattle Populations… Read more »
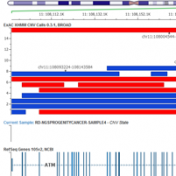
December’s webcast will provide the Golden Helix community with a more in-depth look at CNV analysis in VarSeq. On December 7th, Dr. Nathan Fortier will discuss the challenges and metrics surrounding CNV detection and then demonstrate VarSeq’s new capability from VCF to clinical report. Wednesday, December 7th @ 12:00 PM, EST Numerous studies have documented the role of Copy Number Variations (CNVs)… Read more »