As the need to educate prospective healthcare professionals in the interpretation of genetic data increases, Dr. Jeffrey Moore at the University of Illinois – Urbana-Champaign is using genetics in his chemistry courses. In doing so, Moore is creating a strong connection between the content in his courses and the underlying principles of health and medicine. Last year, Dr. Moore presented a webcast… Read more »
I’m very glad I had the chance to attend ESHG 2016 in Barcelona and talk to so many people about Golden Helix and our software at our booth. ESHG may be the little sibling in size compared to ASHG, but my impression is that it punches above its weight in terms of advancing human genetics applicability to human health and… Read more »
Using Whole Exome Sequencing in distant relationships to identify cardiomyopathy genes Wednesday, June 8th 12:00 pm EDT Cardiomyopathy (DCM; MIM 115200) are myocardial diseases that are frequently hereditary, yet remain gene-elusive for 60% of affected families. Traditional gene discovery techniques dependent on multigenerational samples are difficult to apply. This is because 1. Disease is often impenetrant in the youngest generation,… Read more »
There used to be much energy expended at conferences, bioinformatics forums and even publications about what was the better strategy for interpreting variants of clinical significance: Rule-based filtering and classification mechanisms or rank-based prioritization through all-encompassing “pathogenicity” scores. Both have shown to be effective. Rule-based systems, as exemplified in this filtering diagram in Baylor’s ground-breaking paper on clinical whole-exome sequencing… Read more »
During the webcast yesterday, I demonstrated a few ways of customizing de Novo Candidate and Compound Heterozygous Candidate workflows to consider family structure that was slightly different from the default trio workflow. The families included additional affected and unaffected siblings added to a trio as well as looking at what could be done if there were only two affected siblings… Read more »
Last week, I attended the Advances in Medical Genetics conference in Riyadh, Saudi Arabia. I was asked to present on “Big Data in DNA Analytics”. The event was hosted by Prof. Dr. Majid Alfadhel of the King Saud Bin Abdulaziz University for Health Sciences in collaboration with the Postgraduate Training Center. The event was held to discuss the pros and cons surrounding the… Read more »
Solving the Eigenvalue Decomposition Problem for Large Sample Sizes Since our introduction of the mixed model methods in SVS, along with GBLUP, we have been very pleased to see it used by a number of customers working with human and agri-genomic data. As these customers have grown their genomics programs, the number of samples they have for a given analysis… Read more »
Scott Diehl, a professor and geneticist at Rutgers School of Dental Medicine, performs family studies, association studies and gene mapping to discover the genetic causes of periodontal diseases. Originally, Diehl had a large team of bioinformaticians in the lab to help with his analyses, but the high cost of such personnel created the need for another solution. With Golden Helix’s… Read more »
Submit directly to N-of-One from VarSeq If you or your lab uses N-of-One solutions for clinical annotations, here’s some good news: You can now submit directly to N-of-One from VarSeq! N-of-One’s set of preferred transcripts may differ from those outputted by our algorithms in VarSeq, so our solution was built with that in mind. Our slick, easy to use, and… Read more »
Wednesday, May 11th 12:00 pm EDT VarSeq contains default workflows for Trio analysis which include filter chains for identifying de Novo and Compound Heterozygous variants, but what if you have data for a full Quad or even just a few siblings? How could your VarSeq workflow be adjusted to handle this custom family structure?In this presentation we will demonstrate how… Read more »
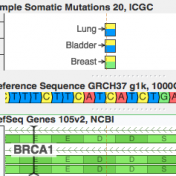
ICGC’s database is now available For quite a while, COSMIC has been synonymous with the catalog of “known somatic mutations”. It is the ClinVar of cancer mutations and invests heavily in “expert curation” (having human experts read papers and pull out references to published somatic mutations). But it turns out there is a new kid on the block, and he… Read more »
New breakthroughs are being made every day in genomics. It’s a dynamic and fascinating industry, and with exceptional growth forecasted in the DNA sequencing market, a new generation of people are entering the field: future researchers, clinicians, counselors and doctors. This new generation will need to learn not only the science, but also understand how to process the massive amounts… Read more »
Clinical reports come in all shapes, sizes and flavors. With that in mind, our clinical reporting interface VSReports was built to be highly customizable and flexible. With a little Javascript and HTML know-how, your clinical reports can be customized to meet the needs and goals of your lab. With a little Javascript and HTML know how, you can customize yours as… Read more »
Dr. Folefac Aminkeng, first place winner in this year’s Abstract Challenge, will present his work to the community via a live webcast on Wednesday, April 20th at noon EDT. Dr. Aminkeng is a Postdoctoral Fellow at The Centre for Molecular Medicine and Therapeutics (CMMT) at the University of British Columbia in Vancouver, BC. Aminkeng’s research focus is the pharmacogenomics of adverse… Read more »
Golden Helix Featured in 20 Most Promising Biotech Technology Solution Providers for 2016! This month, CIO Review released a Biotech Technology Special in which Golden Helix was named among the 20 most promising biotech technology solution providers for 2016. Read the interview with our CEO, Andreas Scherer here: Empowering Precision Medicine through Next-Gen Technology Our customers are monumental in this… Read more »
Did you know you can analyze your Genotyping by Sequencing (GBS) data in SVS? Well you can! You can combine tools for both GWAS quality control and analysis with tools for NGS data analysis to either identify SNPs in your dataset or to identify differences between populations or sub-species. If your species has a reference sequence or even if you… Read more »
Wednesday, April 6th 12:00 pm EDT As the number of samples and associated data volume in a testing lab increases, it becomes imperative for labs to leverage state of the art warehousing technology that not only organizes data, but also aides and enables researchers and clinicians to perform further analysis, and ongoing research. Built on the algorithms and high-performance storage… Read more »
In 1914, the German cytologist Theodor Boveri coined the phrase “Cancer is a disease of the genome”. At this time, his ideas were as equally revolutionary as they were highly contested. Fast forward. More than a hundred years later, Next-Generation Sequencing effectively permits a highly sensitive analysis of cancer cells. It can help us to understand mutations associated with cancer… Read more »
Thank you to all who submitted an abstract in the 2016 Abstract Challenge, it was a great success! With more than 30 submissions and a wide variety of topics, selecting only 3 winners very difficult! With that being said, here are the top 3 winners for this year’s Abstract Challenge: Our 1st place winner is Dr. Folefac Aminkeng, Postdoctoral Fellow… Read more »
Wednesday, March 2nd 12:00 pm EST Clinical labs must have the ability to go from a collection of samples and associated variants to a professional report documenting a short list of clinically relevant variants. Cancer Gene Panels are a common clinical application for genetic tests. In this webcast we will show how VarSeq and VSReports can be used to go… Read more »