Wednesday, September 21st @ 12:00 pm EDT Every day, the trove of genomic data is growing. Clinics are sequencing targeted genes at high read depths to report out genetic tests. Research groups are adding new exomes and genomes to their disease-specific cohorts. Agricultural breeders are genotyping their herds and flocks by the thousands of thousands. The conventional attitude to big… Read more »
After the Wet Lab process has been completed, the bioinformatics analysis of the sequencing data work begins. The next three blogs will focus on three aspects of this process. The building blocks of a bioinformatics pipeline, documentation and validation (today’s topic) Quality Management Clinical Reporting The Building Blocks of an NGS PipelineThe bioinformatics process to analyze NGS data occurs in three… Read more »
Getting the NGS wet bench process right is not a small undertaking. Targeted NGS assays such as multigene panels or exome sequencing allow for the targeted analysis of genomic regions that are of particular interest. For every sample type, e.g. blood, formalin-fixed paraffin-embedded specimens, saliva etc, there must be a detailed protocol in place outlining how each sample type is going… Read more »
For many of you this might be a busy week with school starting up again! However, even though school is just starting, many of our customers have been publishing all summer long. Here are a few highlights from our most recent publications this August: Evaline Ibeagha-Awemu and Xin Zhao of Agriculture and Agri-Food Canada, along with colleagues, published High density genome wide… Read more »
We have come a long way since Next-Generation Sequencing (NGS) evolved as a set of technologies in the 1970s. The higher throughput and rapid reduction of costs associated with NGS have lead to the accelerated adoption of clinical testing that we are experiencing today. Currently, it is applied to analyze inherited diseases, tumors, hematologic malignancies and infectious diseases. It is… Read more »
Join us for a guest presentation Personalized Medicine through Tumor Sequencing by Dr. Jeffrey Rosenfeld! Wednesday, September 7th 12:00 pm EDT The identification of medications that target specific gene mutations is one of the major recent advances in cancer therapy. In 2001 Gleevec was approved to treat patients with the BCR-ABL fusion in chronic myelogenous leukemia (CML). Since then many more drugs… Read more »
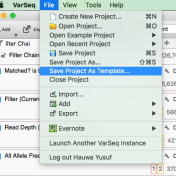
Question: Now that I’ve added annotation sources for my sample, filtered down to a list of interesting variants, flagged those variants and generated a clinical report, can I save or copy the annotation sources and filters for use on another sample? Short Answer: Yes! Long Answer: VarSeq was created with ease and efficiency in mind. In VarSeq, once you’ve defined… Read more »
The adoption of genetic services is key to our ability to provide personalized medicine in the future. The goal is to better diagnose diseases, predict their outcome, and choose the best possible care option for a patient. We still have a long way to go to achieve this goal. While there is agreement about the ultimate goal, there is still… Read more »
Dr. Rohan Palmer presents: Investigating Shared Additive Genetic Variation for Alcohol Dependence Wednesday, August 10th @ 12pm EDT Abstract: Molecular genetic research has supported the use of a multivariate phenotype representing alcohol dependence in studies of genetic association. One recent study found that additive genetic effects on Diagnostic and Statistical Manual of Mental Disorder version four (DSM-IV) alcohol dependence criteria overlap,… Read more »
Now available in SVS! Increasingly important in the analysis of the genotype to phenotype relationship is accurately accounting for the relatedness of samples. This is especially important to model correctly in plant and animal populations where man-directed breeding shapes the relationship structure. Along with trait association, one of the high-value use cases for genotyping animals and plants is to estimate… Read more »
It’s that time of the month again! As usual, we have several new customer publications we would like to recognize and share with you. Please check out a few of our favorite articles: In Biological Psychiatry, Ariel Martinez and Marucio Arcos-Burgos of the National Institute of Health, as well as colleagues, published An Ultraconserved Brain-specific Enhancer within ADGRL3 (LPHN3) Underpins… Read more »
We are excited to announce that earlier this month an article by our CEO, Dr. Andreas Scherer, was featured in Chronicle Pharmabiz. The article is focused on how Precision Medicine is becoming a reality by leveraging the advances in technology. The excerpt below is courtesy of Chronicle Pharmabiz. Almost 2,500 years ago, Hippocrates captured one of the key principles underlying… Read more »
Every month hundreds of clinicians and researchers access the variety of free resources on the Golden Helix website. Our resource library hosts eBooks, webcasts and tutorials to keep the community apprised of new methods, informed on best practices and to help our customers get the most out of their software purchase. Here is a list of the 5 most watched webcasts… Read more »
ClinVar is one of our most used annotations sources for a variety of workflows. It is also the public annotation source that is updated most frequently of all the sources currently supported in VarSeq. ClinVar provides new versions of their database once a month in several formats (XML, VCF, TXT). We use custom Python scripts to convert the provided VCF… Read more »
Since 1999, Bonei Olam has been providing large-scale funding for fertility treatment and research. The non-profit’s mission is to provide whatever means or resources necessary to help childless couples achieve the dream of parenthood. Today, it is recognized in the worldwide medical arena for its leadership role at the forefront of reproductive medicine, research and technology. Specifically, Bonei Olam has… Read more »
Variant Normalization: Underappreciated Critical Infrastructure It may surprise you to learn that every variant in the human genome has an infinite number of representations! Of course, although true, I’m being a bit hyperbolic to prove a point. Even seemingly simple mutations like single letter substitutions are legitimately represented differently in the local context of other mutations that can be described… Read more »
Upcoming Golden Helix Webcast: Using Clinical Reports as part of a Gene Panel Pipeline Wednesday, July 13th @ 12:00 pm EDT VarSeq Reports can be used as part of an automatic pipeline to quickly list variants with information that can be used to make actionable clinical decisions in a readable HTML format. Need to further filter the variants or add interpretation… Read more »
Hearing loss is the most common sensory defect in humans. It affects roughly 1 in 500 newborns, and by the age of 80 approximately 50% of people have some type of hearing loss. Hearing loss has become an enormous burden in healthcare. Perhaps more importantly, studies have shown that hearing loss also affects one’s quality of life, lowering social interactions… Read more »
As spring ends and summer begins here in Montana, we wanted to share another round of customer publications. It’s so important to us that our software plays a role in our client’s success, and we love to display the fruit of their hard efforts each month. Here are a few of the publication highlights: Bradley Aouizerat of UCSF and colleagues published… Read more »
The Bioinformatics Program at Rutgers Cancer Institute of New Jersey, the state’s only National Cancer Institute-designated Comprehensive Cancer Center, plays an integral part in the center’s precision medicine program helping to bring personalized medicine to patients in a timely manner. The Program needs to determine what mutations from a tumor are relevant to a particular therapeutic option, bringing the right… Read more »