Simplifying Custom PGx Annotations with VSWarehouse

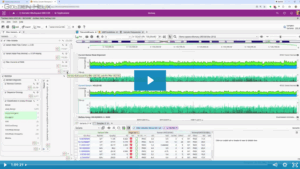
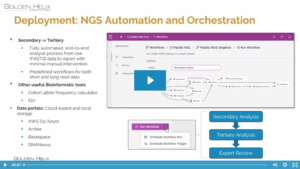
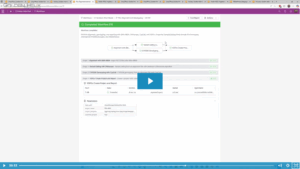
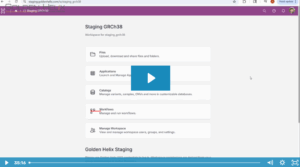
In the rapidly evolving field of pharmacogenomics (PGx), laboratories often face a common challenge: balancing standardized guidelines with the need for specialized, lab-specific data. In the past, maintaining custom PGx annotations was difficult and time-consuming, with custom annotation tracks generated using bespoke curation scripts that merged internal data with VarSeq’s…
Read more →