The current reduced cost and increase availability of genome sequencing has been making academics, clinicians and individuals alike excited with the possibility of increased research depth, diagnosing capability and personal curiosity. And although a freshly sequenced genome is chock-full of tasty letter snippets, the real revelation and education occurs when comparing to an annotation foundation. In this post, I’ll review… Read more »
Ever since the MacArther Lab announced the new gnomAD browser at last year’s ASHG conference, we have had many requests from our customers to make this new variant frequency source available within both VarSeq and SVS. This new dataset includes variants obtained from 123,136 exome sequences and 15,496 whole-genome sequences. In comparison to the original ExAC dataset which contained exomes… Read more »
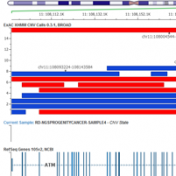
ExAC CNVs were released publicly with a recent publication, providing the full set of rare CNVs called on ~60K human exomes. While there are many public CNV databases out there, this is the first one that was derived from exome data, and thus includes both extremely rare and very small CNV events. With the recent release of Golden Helix’s CNV calling… Read more »
With the release of our updated GWAS E-book, we have recently updated the GWAS example project (SNP Genome-Wide Association Tutorial – Complete). This updated project includes more details about how spreadsheets were generated, how to generate plots and which images were used for the GWAS E-book. This information can be found in the User Notes view in the project navigator and… Read more »
Probably one our most popular public annotation sources we curate and update is the database of Non-Synonymous Functional Predictions (dbNSFP). In it’s recent update, it has expanded the predictions to include FATHMM-MKL and VarSeq now incorporates this new prediction into its voting algorithm of now 6 different discrete predictions per variant. You can update to dbNSFP 3.0 using the built-in… Read more »
I was definitely an early adopter when it comes to personal genomics. In a recent email to their customer base announcing their one millionth customer, they revealed that I was customer #44,299. And I have been consistently impressed with the product 23andMe provides through their web interface to make your hundreds of thousands of genotyped SNPs accessible and useful. It… Read more »
This last week I had the pleasure of attending the fourth annual Clinical Genome Conference (TCGC) in Japantown, San Francisco and kicking off the conference by teaching a short course on Personal Genomics Variant Analysis and Interpretation. Some highlights of the conference from my perspective: Talking about clinical genomics is no longer a wonder-fest of individual case studies, but a… Read more »
In a previous blog post, I demonstrated using VarSeq to directly analyze the whole genomes of 17 supercentenarians. Since then, I have been working with the variant set from these long-lived genomes to prepare a public data track useful for annotation and filtering. Well, we just published the track last week, and I’m excited to share some of the details… Read more »