Our eBook series is made-up of short reads that cover a range of genomic industry topics. From simple bioinformatic concepts to the detailed structure of a genetic data warehouse, we have committed ourselves to provide the community with premium educational resources – at no cost! The series is constantly changing with revisions and new additions, but we’ve found our top… Read more »
Golden Helix is fueled by our customers’ success; every product, every feature, every line of code is created to aid our users and their quest for discovery in the scientific community. As of today, our products have assisted users with 1,200+ publications. A process we know to be long, difficult and incredibly rewarding, it’s an absolute pleasure to be cited… Read more »
Awarded one of the top biotech companies Insight Success recently published its annual Pharma and Life Special Edition announcing the 10 most innovative solution providers of 2018. We are incredibly honored to have received this award and being recognized amongst the top biotech companies! You can access the publication featuring my interview with the editor here: http://www.insightssuccess.com/golden-helix-helping-researchers-clinicians-understand-role-cnvs-human-health-disease/. I have outlined some… Read more »
There are many good reasons why the pursuit of the highest quality genomic interpretation would lead you to the latest human reference. It is more complete and fixes incorrect or partially missing genes that have known implications for human disease. While most major projects cataloging human populations have plans to re-do all their genomic alignments to the new human reference… Read more »
Interpretation of variants in accordance with the ACMG guidelines requires that variants near canonical splice boundaries be evaluated for their potential to disrupt gene splicing [1]. The five most common tools for splice site detection are NNSplice, MaxEntScan, GeneSplicer, HumanSplicingFinder, and SpliceSiteFinder-like. Because these algorithms have been made easily accessible in the bioinformatics tool Alamut, they have been canonized for… Read more »
2017 was an incredibly prosperous year for Golden Helix; we released a handful of new features, announced new partnerships and completed our end-to-end architecture for clinical testing labs. Our webcast series has become a very popular way for our community to stay up-to-date with our new capabilities and best practices in genetic analysis using our software. We had three webcast… Read more »
One of our main focuses in 2017 was VS-CNV which allows clinicians to directly call CNVs in target regions quicker, easier and more affordably than CMA or MLPA testing. Our clients at Robarts Research Institute shared their recent publication with me which confirms that our time and dedication to our CNV capabilities was well worth it. I am delighted to… Read more »
The Golden Helix SNP & Variation Suite (SVS) platform is a powerful and versatile set of tools and algorithms for performing genomic research. That research spans from data originating on genotype micro-arrays to next-generation sequencing. While the majority of SVS users start with genotype data on their samples, any genomic information across a cohort can be used in our various… Read more »
First of all, I wish you a prosperous 2018 along with happiness and health for you and your loved ones. This next year comes with lots of anticipation. We at Golden Helix are looking forward to another year of growth and innovation. Over the last few years, we were able to build a large following of clients in the clinical space…. Read more »
There’s nothing better than starting the New Year off with a great genetics conference in San Diego, CA… We’re looking forward to seeing you all at the Plant & Animal Genome Conference XXVI (PAG 2018)! This year the Golden Helix Team will be represented by Field Application Scientist, Darby Kammeraad, and myself, Content Marketing Manager, Delaina Hawkins. Make sure to… Read more »
We have a lot to thank the 1000 Genomes project for in the genomics community. By the collaborate efforts of many researchers and organizations, the project produced not only the first catalog of rare human variation but in the process standardized many things we take for granted, such as the VCF and BAM file formats. The variant frequencies of the… Read more »
Yesterday’s webcast, Genomic Prediction Methods in SVS, gave attendees a chance to see how the principles of genomic prediction are applied within SVS, predicting phenotypes for both plant and animal species. You can find a recording of the webcast on our site here should you be interested in checking it out or sharing with a colleague! The webcast garnered a… Read more »
It is a great honor for us to be recognized as one of the “Top 25 Biotechnology Solution Providers of 2017” by CIO Applications. You can find my interview with the editor here. We are incredibly thankful for the hundreds of organizations and thousands of users globally trusting our brand! Thank you for your support.
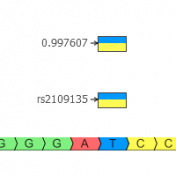
Golden Helix is excited to announce a new round of novel and updated annotations; including a frequency track, a region track, and a gene track. All three of these tracks were created with the use of VarSeq and its Convert Wizard functionality. First, the expansive 1000 genomes track (1kG) has been updated to include sub-population allele frequencies and heterozygous and… Read more »
Golden Helix strives to enable precision medicine by developing powerful software to support researchers with complex analysis. One of my favorite events of the year is our abstract competition. This competition allows us to help our community by recognizing innovative ways to conduct a genomic analysis. I am pleased to announce that our 2017-18 Abstract Competition has officially begun! We would… Read more »
With the recent release of VarSeq 1.4.7, we have expanded the concepts of our popular assessment catalog to include CNV and other region-based records and not just variants. To match these capabilities, we have made a major update to VSWarehouse that supports these new record types in the centrally hosted and versioned Catalogs and Reports. Review of the VSWarehouse Genomic… Read more »
With the recent upgrade to VarSeq 1.4.7, users gain access to some new great features. Among the additions are new CNV annotations (Figure 1). In this final chapter of the annotation blog series, we are going to provide descriptions of the new CNV annotations and how they can be used. The types of CNV annotations vary and include frequency, clinical… Read more »
We’ve returned from another successful year at ASHG and had an incredible time. I find the conference to be a terrific opportunity to connect with our customers, partners, and friends in the industry. This year’s Presidential Symposium was a true delight featuring a discussion of global health and genomics between two absolute legends in the health and science world: Bill Gates,… Read more »
In our final chapter of this variant annotation blog series, we will discuss additional annotations that provide powerful variant filtering and analysis capability. Golden Helix curates many annotations in a way that allows for simple analysis and saves the users the hassle of all this data management. Whether you are trying to capture rare variants known across multiple subpopulations in… Read more »
VarSeq Updated with CNV Annotations, CNV PhoRank, and Region Assessment Catalogs This year we have released multiple advances to support CNV analysis in VarSeq, expanding our target region based VS-CNV caller to handle exomes and low-depth genomes as well as additional supporting algorithms like calling Loss of Heterozygosity regions. To top this off, VarSeq 1.4.7 has been shipped with many… Read more »