Whole exome sequencing workflows using SNP & Variation Suite (SVS) was presented in a recent webcast, by Dr. Robert Hamilton from the Hospital for Sick Kids. In particular, he performed some filtering on his data to look for only heterozygous variants in his sample of interest, removed variants with allele frequency less than 0.005% based off of the ExAC Variant Frequency… Read more »
When the new human reference genome was released over two years ago, it was hailed as a significant step forward for next generation sequencing. Compared to GRCh37, the new GRCH38 reference assembly fixed gaps, repaired incorrect sequences and offered access to sections of the genome that had been previously unaccounted for. Despite these improvements, adoption of the new assembly has… Read more »
The power of VSPipeline is in it’s ability to automate VarSeq workflows. Using VarSeq to create a pipeline template is great because it allows you to dial in the applied filters as well as interactively organize the annotations and applied algorithms. Automating a workflow with VSPipeline is straightforward when beginning with an existing project. However, there are several steps that… Read more »
There used to be much energy expended at conferences, bioinformatics forums and even publications about what was the better strategy for interpreting variants of clinical significance: Rule-based filtering and classification mechanisms or rank-based prioritization through all-encompassing “pathogenicity” scores. Both have shown to be effective. Rule-based systems, as exemplified in this filtering diagram in Baylor’s ground-breaking paper on clinical whole-exome sequencing… Read more »
Submit directly to N-of-One from VarSeq If you or your lab uses N-of-One solutions for clinical annotations, here’s some good news: You can now submit directly to N-of-One from VarSeq! N-of-One’s set of preferred transcripts may differ from those outputted by our algorithms in VarSeq, so our solution was built with that in mind. Our slick, easy to use, and… Read more »
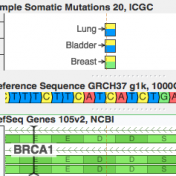
ICGC’s database is now available For quite a while, COSMIC has been synonymous with the catalog of “known somatic mutations”. It is the ClinVar of cancer mutations and invests heavily in “expert curation” (having human experts read papers and pull out references to published somatic mutations). But it turns out there is a new kid on the block, and he… Read more »
Did you know you can analyze your Genotyping by Sequencing (GBS) data in SVS? Well you can! You can combine tools for both GWAS quality control and analysis with tools for NGS data analysis to either identify SNPs in your dataset or to identify differences between populations or sub-species. If your species has a reference sequence or even if you… Read more »
Wednesday, April 6th 12:00 pm EDT As the number of samples and associated data volume in a testing lab increases, it becomes imperative for labs to leverage state of the art warehousing technology that not only organizes data, but also aides and enables researchers and clinicians to perform further analysis, and ongoing research. Built on the algorithms and high-performance storage… Read more »
As VarSeq continues its adoption amongst clinical labs and researchers looking for reproducible workflows for variant annotation, filtering and interpretation, we have continued to prioritize the addition of features to assess the quality of the upstream data at a variant, coverage and now sample level. The Importance of Quality Assurance Sample prep and sequencing problems are difficult to detect through the analysis… Read more »
Scaling is in our DNA: Making Genomics Accessible One of the things I absolutely love about the work we do at Golden Helix is keeping up with the changes in data analysis driven by the iterative and generational leaps in technology. But one thing has always been a constant since day one: we break preconceived notions of what scale of… Read more »
Yesterday, it was my pleasure to share in a live webcast our integrated solution for genetic data warehousing, VSWarehouse. Although we had a great set of questions at the end of our presentation, we didn’t have time to answer all of them, so here is a selection of the remaining representative questions and my answers. We can provide a hosted version… Read more »
Compound Heterozygous Workflows: Including a 2nd Affected Child Looking for Compound Heterozygous regions for a trio is fairly straight forward in VarSeq, we include this workflow in our shipped Exome Trio Template. An example of which is included with our Example Projects which can be found by going to File > Example Projects > Example YRI Exome Trio Analysis. But… Read more »
There is no doubt that we have big data in the field of genomics in general and Next Generation Sequencing specifically. Illumina’s latest HiSeq X can produce 16 genomes per run, resulting in terabytes of raw data to crunch through. Yet all that crunching is not the hard part. So, what is the main obstacle to scientists being able to… Read more »
Golden Helix in 2016 We had a terrific year 2015. It was the year in which we got serious about the clinical testing market. We successfully continued on the path of attracting more referenceable clients such as University of Iowa, Baby Genes, Prevention Genetics and many more. We rounded out our VarSeq suite by adding more clinically relevant features and… Read more »
The most common use of the VarSeq Match Gene List algorithm of course is to determine if the variants in your data set are contained within your genes of interest. As an example of this, say you are working with a whole exome trio and only want to consider those variants that are contained within the 56 genes recommended by… Read more »
As VarSeq’s adoption has grown among analysts using whole exome data to diagnose rare diseases, a couple of family designs outside of the common trio of an affected child and both parents have come up frequently. While having both parents provides the maximum power to discover de novo mutations and recessively inherited variants, it is not always possible to contact… Read more »
With the release of VarSeq 1.3.1 we have included a new demo project to showcase a single tumor-normal pair analysis workflow. The project can be accessed through VarSeq and VarSeq Viewer by going to File > Open Example Projects > Example Tumor-Normal Pair Analysis. This project contains an exome pair (Normal-N990005 and Tumor-T990005) from the Gastric Cancer study Exome sequencing of… Read more »
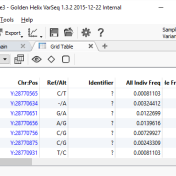
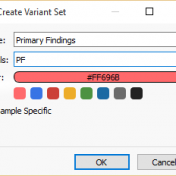
With the release of VSReports, we added the ability to “select” rows of your filtered output (often variants, but potentially things like coverage regions or genes) with a new feature dubbed “Record Sets”, but more often described as “colored checkboxes” for your tables. Although necessary for the important task of marking primary, secondary or other sets of variants for a… Read more »
As VarSeq gains in popularity, we want to give Viewers and customers alike the opportunity to look at projects that are completed from start to finish. To this end, VarSeq (and VarSeq Viewer!) currently comes with two demonstration projects, Example TruSight Cardio Gene Panel and Example YRI Exome Trio Analysis. To access these projects from the VarSeq start page go to… Read more »
Our webcast yesterday featured two clinical workflows and and the ease in moving from an unfiltered variant file to a clinical report containing the variants of interest using VarSeq and VSReports. There were several great questions and I wanted to pass on a few of particular interest. Question: Are annotation sources included in VarSeq for free?