Recent Customer Publications We are halfway through November, and it’s hard to believe how fast the holidays are approaching. Today, we wanted to recognize recent customer publications all over the world, from China to Australia and back to the United States. Congratulations to all who published, and we hope to see many more through the end of the year! Mark… Read more »
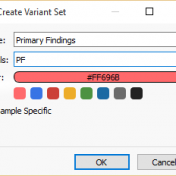
With the release of VSReports, we added the ability to “select” rows of your filtered output (often variants, but potentially things like coverage regions or genes) with a new feature dubbed “Record Sets”, but more often described as “colored checkboxes” for your tables. Although necessary for the important task of marking primary, secondary or other sets of variants for a… Read more »
As VarSeq gains in popularity, we want to give Viewers and customers alike the opportunity to look at projects that are completed from start to finish. To this end, VarSeq (and VarSeq Viewer!) currently comes with two demonstration projects, Example TruSight Cardio Gene Panel and Example YRI Exome Trio Analysis. To access these projects from the VarSeq start page go to… Read more »
Give our SVS viewer a try today! Interested in seeing what the SNP & Variation Suite (SVS) software can do? Download the free SVS Viewer! With the SVS Viewer, you can explore and interact with the workflows of a pre-built projects. To get you started, we have included a SNP GWAS project for you to download. And don’t worry, it is… Read more »
Our webcast yesterday featured two clinical workflows and and the ease in moving from an unfiltered variant file to a clinical report containing the variants of interest using VarSeq and VSReports. There were several great questions and I wanted to pass on a few of particular interest. Question: Are annotation sources included in VarSeq for free?
Bipartisan Budget Act of 2015 Passed! We at Golden Helix, and I are certain that many in our community, are more than pleased to hear that President Obama has signed the “Bipartisan Budget Act of 2015“. Most importantly, the budget agreement raises discretionary spending by $80 billion in fiscal years 2016 and 2017 which should create room for increased spending… Read more »
After our announcement in August that we would be making GxE Regression available in SVS, we were pleased to receive feedback that this was exactly what our customers had been wanting. Being able to account for environmental effects or gene effects as interacting with the SNPs was essential to those researchers working with GWAS. Unfortunately, this did not help our customers who were… Read more »
What you need to know At the beginning of October, VSReports came to VarSeq. Since then, we have been getting rave reviews from our customers. As Dr. Andreas Scherer mentioned earlier this month, VSReports takes the output of your tertiary variant analysis and converts it into a fully customized and actionable clinical grade report in just one click. Additionally, VSReports ships with… Read more »
While VarSeq comes with a number of starter workflows that are stored as templates, customers also have the option of creating filter chains from scratch; analyzing a single exome may require you to do exactly that. In this blog, I’ll go through analyzing a single exome and generating a list of variants for further study. After importing the variant data… Read more »
The month of October is flying by, and we have already had some great publications by some of our customers here at Golden Helix. We wanted to share them with you, and hope you find them as interesting as we did. Congrats to all who published! Bahram Namjou of the Cincinnati Children’s Hospital and colleagues published A GWAS Study on… Read more »
ASHG 2015 in Baltimore was a very good conference for us at Golden Helix. It was a delight to connect with existing clients as well as interested members of our community. The conference is a premium networking opportunity for professionals in our field, with an ever increasing number of talks, demos and a constantly expanding vendor exhibition area. … Read more »
Our final webcast presentation in the series of winners from our Annual Abstract Challenge is first place recipient Dr. Sergey Kornilov. A Postdoctoral Associate in the Child Study Center at Yale University’s School of Medicine, Dr. Kornilov’s submission focused on the genetic basis of developmental language disorders in a geographically isolated Russian-speaking population. Next week on October 14th, he will… Read more »
While stand-alone SVS is an amazing statistical software package for genomics analysis, adding additional Python scripts to the program can expand SVS’s genomic analysis capabilities. In this post, I’ll take you through the most frequently downloaded Add-On Scripts for SVS and the top five were not what I expected! Coming in at Number Five is the script Convert Dosages to… Read more »
The Golden Helix® team is gearing up for ASHG 2015 in Baltimore as I would guess many of you are too. I for one am super excited since I grew up in the Baltimore area and have a fondness for Inner Harbor and crabs! This year, you will find Ashley Hintz (our Field Application Scientist) and myself just inside the… Read more »
The 65th annual ASHG in Baltimore will be another exciting one. We at Golden Helix have been very busy this year making great improvements to both SVS and the VarSeq software and we look forward to showcasing them during our in-booth demos. In particular, we will launch two new additions to the VarSeq software suite; VSReports and VSWarehouse. VSReports brings highly customizable clinical… Read more »
Yesterday, our VP of Product and Engineering, Gabe Rudy, presented VSReports to the Golden Helix community for the first time in a live webcast; Authoring Clinical Reports in VarSeq. It was an excellent presentation. Gabe highlighted VSReports’ ability to take the output of tertiary analysis to a customized clinical grade report in one click. He also gave an overview of… Read more »
This month we wanted to highlight four publications from a few of our valued customers. Congrats to all! Zuemei Ji and Christopher Amos of Dartmouth University published The Role of Haplotype in 15q25.1 Locus in Lung Cancer Risk: Results of Scanning chromosome 15 in the Oxford Journals which analyzed lung cancer patients and a control study to determine that rs588765-rs16969968 may… Read more »
The next release of VarSeq will ship a new product that is highly relevant to our customers in clinical testing labs. Via VSReports, VarSeq now has the ability to generate clinical-grade reports. These reports are fully customizable, containing focused and actionable data. VS Reports ships with report templates that are modeled off of the ACMG guidelines, the de-facto gold standard… Read more »
Precision Medicine e-Book “It’s far more important to know what person the disease has than what disease the person has.” – Hippocrates (460 BC – 370 BC) Almost 2,500 years ago, Hippocrates captured one of the key principles underlying precision medicine. In the 21st century we take the understanding of the individual characteristics of a person to a new level…. Read more »
With our latest release of VarSeq last Thursday, we are proud to offer the VarSeq ® Viewer to the community, for free! When you download VarSeq ® Viewer, you can explore pre-built projects to interact with the annotations and filters the project provides, visualize selected variants with pile-ups, and export data to widely used formats. To get you started, we have included… Read more »