Hypertrophic Cardiomyopathy History It was December 9th, 1989, when one of Loyola Marymount’s strongest inside players, Hank Gathers, collapsed during the middle of a collegiate level basketball game against UC Santa Barbara. Measuring in at 6’7” and weighing 210 pounds, Gathers was diagnosed with exercise-induced ventricular tachycardia, or in layman’s terms, an abnormal heartbeat. Even with the concerning nature of… Read more »
Overview VSClinical enables users to evaluate variants according to the ACMG guidelines in a high-throughput fashion and obtain consistent results and accurate variant interpretations. This feature is tightly integrated into our VarSeq platform as well, and when paired together, users can evaluate NGS data and obtain clinical reports all in one suite. Coupled with the ability to find novel or… Read more »
VSClinical provides a rapid-fire way to investigate any variant’s impact by following the ACMG Guidelines process for classification. We will be demonstrating this by looking at interesting examples of rare disorders and showcasing some evaluation steps users may deploy in their analysis. Our first example in this blog series is for a patient who has an indifference to pain, while… Read more »
Golden Helix, Inc. has announced themselves as the recipient of an NIH SBIR Grant 2R44GM128485-02 entitled “Automated and Guided Workflows for Clinical Testing Using NGS Assays.” “With the help of this grant, we will take the clinical automation of NGS data to the next level, focusing on germline diseases and cancer diagnostics,” states Andreas Scherer, Ph.D., President, and CEO of… Read more »
The Department of Clinical Genetics at Odense University Hospital offers a variety of genetic analyses for families of syndromic children and other inherited conditions, averaging 4,000 genetic analyses per year. In 2016, the lab decided to introduce whole exome sequencing to their offerings to take over a lot of the work they were currently conducting via gene panel analysis. They… Read more »
Happy New Year. I trust you had a relaxing time over the holidays with family and friends as well as a great start into 2019. Golden Helix certainly had a landmark year. There were many highlights that actually shaped the direction for us in the years to come. Please let me mention a few: Essentially, we had a blast in… Read more »
VSClinical is our most recent product that allows users to evaluate variants according to the ACMG guidelines. As with any tertiary analysis, there is a need to implement best practices into your workflow and using VSClinical for the ACMG guidelines is no exception. That said, we have put together a Best Practices Blog Series, with the purpose of discussing some… Read more »
We just got back from three busy days at the Molecular Pathology (AMP) conference in friendly San Antonio, Texas. Keeping up the Golden Helix conference momentum for the year, we had 3-4 in-booth demonstrations a day covering our CNV calling, variant interpretation, and data warehousing products for NGS-based genetic tests. And in short, NGS based tests for cancer and germline… Read more »
This webcast generated some great questions! If you have any other questions for me that are not answered below, please feel free to ask those by emailing [email protected]. Does VSClinical come with support for the new reference genome? Yes! We worked hard to make everything work in VSClinical regardless of your choice of reference genomes. The only caveat is that… Read more »
In my recent webcast, I demonstrated how VS-CNV users can detect high-quality copy number variant events. If you didn’t have a chance to join, you can view the recording below! This webcast generated a lot of great questions! If you have any other questions about the content covered in this webcast that is not answered below, please feel free to… Read more »
Genome-wide association studies (GWAS) are useful in genetics as they test for the association of a phenotype with common genetic variants. GWAS is “hypothesis-free” and does not require prior knowledge of a gene’s biological impact on a trait. The catch though is that this leads to analyzing hundreds to thousands of genome-wide array samples to elucidate single nucleotide polymorphisms (SNPs) associated with a specific phenotype.
It’s fascinating to hear the various ways our products are being used by customers all around the world. This month we’re featuring publications citing VarSeq, SVS and HelixTree which cover studies from Schizophrenia, Multiple Sclerosis and more. We hope you enjoy! Exome sequencing in schizophrenic patients with high levels of homozygosity identifies novel and extremely rare mutations in the GABA/glutamatergic pathways This… Read more »
We don’t just like hearing what our clients are up to … we love bragging about what they’re doing to the world as well! This week we’re showcasing Dr. Stanley Nelson, and his team at UCLA Health, who used next-generation sequencing and our VarSeq software to help diagnose a child’s long running medical mystery. Audrey Lapidus knew there was something… Read more »
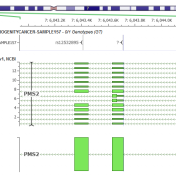
The support team at Golden Helix is always here to help with your SVS and VarSeq needs. Often, we receive some excellent questions that should be shared with the rest of our users. This blog will answer some common questions we’ve been seeing lately regarding VarSeq CNV. I’ve noticed there is a version 2 of the CNV caller on Targeted Regions Algorithm, how has… Read more »
Human genetic variation makes us unique. On average, humans are to 99.9% similar to each other. Understanding in detail what the nature of the difference in our genetic make-up is all about allows us to assess health risks, and eventually enables Precision Medicine as we determine treatment choices. Furthermore, it enables scientists to better understand ancient human migrations. It gives… Read more »
Our July webcast presentation will be focused on clinical workflows in VarSeq. We wanted to share the full details with you and hope you are able to attend! Wednesday, July 12th 12:00 PM EST This month’s webcast is a VarSeq exploration, featuring several example workflows and helpful features in VarSeq that can be used in the clinic. We will discuss… Read more »
The current reduced cost and increase availability of genome sequencing has been making academics, clinicians and individuals alike excited with the possibility of increased research depth, diagnosing capability and personal curiosity. And although a freshly sequenced genome is chock-full of tasty letter snippets, the real revelation and education occurs when comparing to an annotation foundation. In this post, I’ll review… Read more »
Today, we are happy to announce a multi-year partnership with Sentieon, a company that develops bioinformatics secondary analysis tools to process genomic data. This partnership will integrate Sentieon’s secondary analysis tools with Golden Helix software to provide users with a comprehensive solution for genomic data analysis. Sentieon’s suite of secondary analysis tools made the significant improvement in runtime over BWA-MEM, GATK,… Read more »
We had lots of customers publish their work using our SVS software, and I wanted to share their work with you. Congrats to all! Here are some of the highlights: Francesca Fernandez of the University of Wollongong along with colleagues published Effects of common GRM5 genetic variants on cognition, hippocampal volume and mGluR5 protein levels in schizophrenia in Brain Imaging and… Read more »
It may be possible to say that annotating a variant correctly and accurately against gene transcripts is the most important job of a variant annotation and interpretation tool. We take it very seriously at Golden Helix as we support VarSeq and its use by our customers in both research and clinical contexts. It has been a source of frustration that… Read more »