In the 1990s the genetic industry voiced a request for a variant catalog that incorporates associated variant information such as phenotypic and metabolic pathways. The call was answered by NCBI, which created dbSNP; dbSNP became publicly available in 1998 and around 1.5 million variants. Fast forward to the present and dbSNP now contains over 2 billion SNPs spanning human, rat,… Read more »
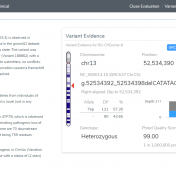
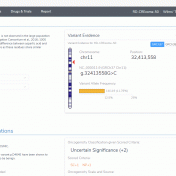
We have now reached the final blog of the NGS-Solutions for Clinical Variant Analysis series. Part I of this series explored the capture of variant classifications in the VSClinical environment when following the ACMG and AMP guidelines. Part II was similar in content but for the capture of clinically relevant copy number variants as well as using a CNV catalog… Read more »
It doesn’t take much effort to find articles discussing the value of Next-Generation Sequencing (NGS). There is a consistent tone amongst authors that implementing NGS pipelines are critical for clinical efficiency in both hereditary disorders and somatic. However, NGS strategies do not come without their own challenges. Challenges include not only the detection and calling of high quality/probability variants from… Read more »
It is common knowledge that variants can be germline or somatic depending on whether the variant was inherited or acquired after birth. A well-known example is cancer-causing mutations in the BRCA genes, wherein the mutation may or may not have been inherited. Understanding the origin of the cancer-causing mutation is important when assessing potential treatment options as well as identifying… Read more »
Introduction: Malignant Rhabdoid tumors (MRT) are among the most aggressive and lethal forms of infant and child cancer (1). These tumors are characterized by an unusual combination of mixed cellular elements similar to but not typical of teratomas and can originate at any anatomic location. When MRTs are present in the brain, they are called atypical teratoid/rhabdoid tumors (AT/RT), which… Read more »
As I prepared to write the Customer Publication blog this month, I was excited by the number of recently published papers that stood as examples of how both VarSeq and SVS software are employed to advance diagnostics and treatments in human medicine. We often think of SVS as the go-to platform for Agrigenomics, however both of our platforms have broad… Read more »
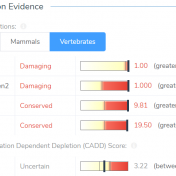
The University of Washington’s Combined Annotation Dependent Depletion (CADD) algorithm measures the deleteriousness of genetic variants. This includes single nucleotide polymorphisms (SNVs) and short insertions and deletions (indels) throughout the human reference genome assembly. This algorithm was introduced in 2014 and has since become one of the most widely used tools to assess human genetic variation. Since 2014, the algorithm has been… Read more »
Abstract Before assessing the clinical significance of a somatic mutation, one must determine if the mutation is likely to be a driver mutation (i.e. a mutation that provides a selective growth advantage, thereby promoting cancer development). To aid clinicians in this process, VSClinical provides an oncogenicity scoring system, which uses a variety of metrics to classify a given somatic mutation… Read more »
An under-appreciated area of complexity when looking into the field of genetics from the outside can be found in genes and transcripts. Alternative splicing allows eukaryotic species to have a wonderfully powerful genetic code, resulting in multiple protein isoforms being encoded in a single section of DNA. But when it comes to variant interpretation, different transcripts can result in widely different predicted… Read more »
Writing this blog post to summarize and highlight our customer’s publications is undoubtedly one of my favorite things to do! The wide variety of topics is always surprising and inspiring, and I am humbled by the efforts of dedicated scientists who are helping to protect and enrich our lives in so many ways. Our SNP & Variation Suite (SVS) software… Read more »
This morning I released a new version of my eBook “Clinical Variant Analysis for Cancer – Second Edition.” The clinical utilization of Next-Generation Sequencing data to diagnose cancer has taken off, resulting in the need to standardize the interpretation and reporting of observed genomic variations. This eBook explores the entire clinical diagnostic process. It demonstrates how Golden Helix software can support clinicians with their… Read more »
Thank you to everyone who joined me for our latest webcast, “Next-Gen Sequencing of the SARS-CoV-2 Virus with Golden Helix.” If you missed the live event and are interested in knowing what we talked about, you may access the recorded event below: Our Live Q&A generated a lot of great questions. Unfortunately, we were unable to answer them all, but… Read more »
Golden Helix has been committed to making sure our community has the tools and resources they need during this pandemic. We have developed several different assistance programs, such as our home office licenses for current users and our software discounts for COVID-19 related projects, to name a few. We have been approached by a number of labs not working with COVID-19… Read more »
It is an honor to be published in The Journal of Precision Medicine’s March 2020 issue where I have outlined the current best practices for NGS-based cancer diagnostics. In this article, I detail: The need to standardize clinical reporting Popular annotation sources and functional prediction algorithms The AMP Guidelines and how these can be applied with Golden Helix’s Diagnostic Platform… Read more »
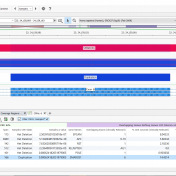
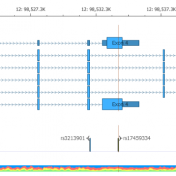
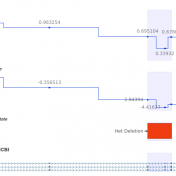
In trio workflows, one of the most important factors in scoring a variant is understanding how that variant is inherited from the parents. Likewise, when looking at extended families, the segregation, or presence of the variant among the affected versus unaffected individuals provides evidence for its pathogenicity for a given phenotype or disease. Given the nature of Copy Number Variants… Read more »
*The following text is only a preview of our new eBook “Genetic Analysis of the COVID-19 Virus and Other Pathogens”. Request a free copy to access the full version.* SARS-CoV-2 has just been recently discovered. The knowledge about this virus is fairly new and certain aspects of it are still under review or in-flux entirely as we learn more about… Read more »
VarSeq 2.2.1 was released on April 1st and features an upgraded gene annotation capability with new RefSeq genes tracks and an AMP workflow addition: the Drugs and Trials tab. The new RefSeq human genome genes tracks contain updated gene names and the recognition of any MANE (Matched Annotation from NCBI and EMBL-EBI) identified transcripts. VarSeq has been updated to be… Read more »
Thank you to everyone who joined me for yesterday’s webcast, I hope you all enjoyed it. If you missed the live event and are interested in knowing what we talked about, good news, you can watch the recorded version right here! There were so many great questions asked during our Live Q&A that I was unable to answer all of… Read more »
*The following text is only a preview of our new eBook “Genetic Analysis of the COVID-19 Virus and Other Pathogens”. Request a free copy to access the full version.* At the end of 2019, a virus appeared somewhere in the Chinese city of Wuhan. It caused cold and flu-like symptoms in most, but also pneumonia and death in a few…. Read more »
Our Support Team curates a variety of tutorials to help orient new users to the capabilities of VarSeq. We are happy to announce the team’s new release of the trio tutorial that places emphasis on using the ACMG guidelines. This tutorial gives insight into the proper setup of pedigree structure as well as detailed descriptions of the filter containers and… Read more »