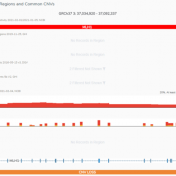
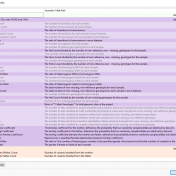
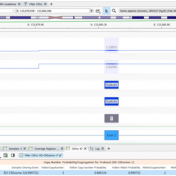
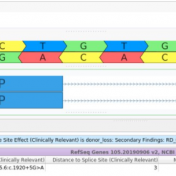
The collaboration between the Clinical Genome Resource (ClinGen) consortium and the American College of Medical Genetics (ACMG) recently developed published guidelines for the interpretation of CNVs called on next-generation sequencing data. These new guidelines are the first to provide a robust set of rules for the interpretation of small intragenic deletions and duplications and are now automated in VSClinical. … Read more »
In many cases, VarSeq users typically run single trio projects or perhaps an extended family project. Not only are all the inheritance model algorithms available in the VarSeq software to capture de novo, dominant, or recessively inherited variants but there are a number of quality control fields to help ensure the pedigree was set up properly. The last thing any… Read more »
The recent release of VarSeq 2.2.2 brings our Word report template system, previously featured in VSClinical AMP, to the VSClinical ACMG workflow. This blog post will describe how to use the Word template system using one of our shipped templates as well as how to start customizing your own templates. We will cover the three different report templates that ship… Read more »
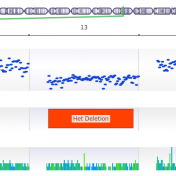
VarSeq 2.2.2 has incorporated a multitude of interesting new features. In this blog, I want to continue discussing these features and how each can be incorporated into your workflow. I will also discuss the application of the Probability Segregation algorithm for copy number variation (CNV) analysis. The Probability Segregation algorithm is a new algorithm that has been added to VarSeq… Read more »
Our latest VarSeq release is one of the largest we’ve ever had, boasting an extensive list of new features and improvements. As part of this release, we have dramatically expanded our support for splice site analysis. This includes improvements to our novel splice site algorithm and support for splice site effect prediction along with several other small improvements. Novel Splice… Read more »
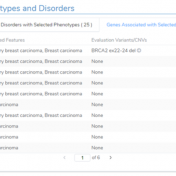
Our recent release of VarSeq 2.2.2 comes with a long list of upgrades and new features. In this blog post, we will demonstrate how defining sample phenotypes are available in VSClinical. One noticeable change is the ACMG guideline variant evaluation in VSClinical. Not only has this interface added CNV guideline evaluation, simplified the reporting process with embedded Microsoft Word and… Read more »
Golden Helix offers a market-leading bioinformatics solution that allows users to evaluate next-generation sequencing variants according to the ACMG and now ACGS guidelines. The ACMG guidelines were created in 2015 and are widely accepted as best practice for the interpretation of sequencing variants throughout the United States (Richard et al 2015). Very similar are the ACGS guidelines that were developed… Read more »
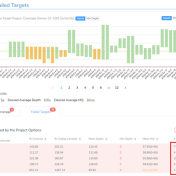
Curious about how coverage statistics can be used in conjunction with VarSeq? Evaluating the coverage over target regions or whole genomes is essential whether you are working with variant or CNV analysis. VarSeq has had the capability to compute sample level coverage statistics for some time now, but in the 2.2.2 release of VarSeq, there are some new features that… Read more »
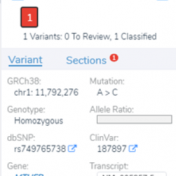
In this blog post, I will be analyzing a loss-of-function splice variant in MTHFR using VarSeq. In the search for clinically relevant variants contributing to rare disorders, efficient filtering strategies are an important step in eliminating disinteresting variants. However, any applied filters must also ensure no interesting variants inadvertently get filtered out. Golden Helix provides the tools to complete this… Read more »
In the webcast, Evaluation of Copy Number Variants with VSClinical’s New ACMG Guideline Workflow, we discussed how VSClinical implements Section 4 of the ACMG guidelines. Specifically, we focused on integrating literature and publications to assess the pathogenicity of a CNV event when there was a lack of dosage sensitivity information. One of the primary pieces of evidence for evaluating genes… Read more »
In a recent webcast, users were exposed to some new features upcoming in the next release of VarSeq. In this update, we will use an example de Novo CNV in a cardiomyopathy panel in VarSeq. There is a long list of new tools and polishes to the software but one major upgrade is the inclusion of the ACMG and ClinGen… Read more »
A common discussion with our customers includes the challenges with the tertiary stage of analyzing next-gen sequencing data. This is the stage where all data from gene panels, exome, or whole genome scale pass through filters to quickly isolate the clinically relevant variant contributing to a patient disorder. Golden Helix has recognized these challenges in the scale of data and… Read more »
Did you know you can control your preferred transcript settings for clinical interpretation in VSClinical? Your lab is analyzing the DNA of a tissue sample from a patient with small cell lung cancer. The lab technician has imported the patient data into VSClinical to detect clinically relevant variants and evaluate and score these according to the AMP Guidelines, as well… Read more »
In this blog update, I’ll be walking you through some of the advanced plotting capabilities with GenomeBrowse. The strategy with any next-gen sequencing analysis is to filter down to interesting variants for either research or clinical conclusion. Golden Helix produces powerful software specifically tailored for this efficient and comprehensive search for interesting and clinically relevant variants. One additional advantage of… Read more »
Our software solutions and partners have brought dramatic improvements to the secondary and tertiary analysis stages of variant evaluation. Regarding secondary analysis, we’ve discussed increased efficiencies in speed and overall accuracy in the variant calling process with Sentieon. On the tertiary side, we have explored numerous workflows in VarSeq highlighting filtration to clinically relevant variants, as well as the automated… Read more »
We have had many customers come to us over the years with a simple problem: they have BAM files for whole exome or gene panel data and would like to call CNVs using VarSeq’s powerful CNV calling capabilities, but they don’t have a bed file defining the target regions for their samples. To address this problem, we have developed a… Read more »
In VarSeq 2.2.1, you can set template annotation sources to automatically update to the latest version. Previously, VarSeq templates were frozen in time. Now, each new project created from a template would use the same source that was used when the template was created. When you save a template, you can have the sources automatically update to the latest version…. Read more »
Our Support Team curates a variety of tutorials to help orient new users to the capabilities of VarSeq. We are happy to announce the team’s new release of the trio tutorial that places emphasis on using the ACMG guidelines. This tutorial gives insight into the proper setup of pedigree structure as well as detailed descriptions of the filter containers and… Read more »
Generating a clinical report is the final step of most NGS pipelines and is important as it relays results and information to legacy systems, physicians and ultimately the patient. As reporting is a valuable process, Golden Helix offers reporting capabilities according to the ACMG and AMP guidelines but also as a standalone feature in VSReports. VSReports is a platform that… Read more »
At Golden Helix, we want our new users to hit the ground running with VarSeq and not spend oodles of time getting started building and automating their workflows. To achieve this goal, our team has generated blogs, webcasts, and tutorials that explain and demonstrate workflows that are possible with VarSeq. Each VarSeq tutorial offers step by step instruction in which… Read more »