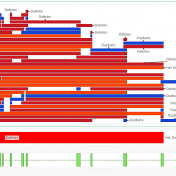
Breakends (BNDs) represent the precise genomic coordinates where DNA is rearranged in structural variants such as translocations, inversions, and complex rearrangements. Analyzing breakends in NGS analysis is important because they capture the exact “breakpoints” of structural alterations, enabling accurate identification of gene fusions, large deletions, or other events that can disrupt or activate genes of clinical relevance. Systematically tracking how… Read more »
We’re thrilled to announce that VarSeq 3.0.0 is right around the corner, and with it comes one of the most exciting updates yet: a complete reimagining of the Assessment Catalogs system. Whether you’re curating clinically relevant variants, tracking tumor-specific calls, or building rich catalogs of sample information, the new system is designed to be more powerful, more versatile, and easier… Read more »
Variant interpretation is an iterative process, and over time, labs can accumulate a large collection of reviewed variants. Managing these interpretations efficiently is critical for saving time, reducing redundancy, and maintaining consistency across projects. VarSeq assessment catalogs serve as a central repository for storing variant classifications, interpretations, and supporting details. They capture key information such as: These catalogs make it… Read more »
Over the past several months, we’ve highlighted the powerful new capabilities introduced with VarSeq Warehouse 3.0. Today, I’d like to focus on a familiar feature that’s been thoughtfully enhanced, assessment catalog record management. This update significantly improves data quality, role-based collaboration, and regulatory compliance. Variant assessment catalogs have always been a core component of the VarSeq platform, allowing users to… Read more »
Variant interpretation is a critical aspect of any clinical NGS workflow. VarSeq assessment catalogs are a tool used to save variants and associated variant information for easy tracking and retrieval of completed variant interpretations. As variant interpretations stack up and classifications are saved, storing in assessment catalogs makes it easy to automatically fill in a previous interpretation if the variant… Read more »
Assessment catalogs are a way for VarSeq users to save variants and variant information for clinically relevant variants, so when you come across this variant again in another sample, all of the work to analyze and classify the variant is already done! But there is more than meets to the eye when it comes to using assessments in VarSeq. Being… Read more »
When doing clinical variant analysis, it is often essential to keep track of the variants that are encountered in each sample, their pathogenic or oncogenic classifications, which individual created or saved an interpretation, and when. For this purpose, VarSeq prompts VSClinical users to create default assessment catalogs in which to store variants and other events. However, we are aware that… Read more »
Discover how to enhance collaboration in VSClinical by sharing variant scores and assessment catalogs through the strategic relocation of the AppData folder. A subtle but powerful utility of VSClinical concerns the ability to share variant scores between users on the same account. What I mean by that is, leveraging the location of the AppData folder, so that users are writing… Read more »
In this blog, we will be covering new assessment catalogs and how they work to improve saving and tracking variant interpretations. VarSeq is a variant analysis tool that effectively analyzes single nucleotide (SNVs) and copy number variants (CNVs) in both cancer and germline workflows. Because VarSeq enables such diverse variant analysis, there are many research labs and institutions that evaluate… Read more »
Our software solutions and partners have brought dramatic improvements to the secondary and tertiary analysis stages of variant evaluation. Regarding secondary analysis, we’ve discussed increased efficiencies in speed and overall accuracy in the variant calling process with Sentieon. On the tertiary side, we have explored numerous workflows in VarSeq highlighting filtration to clinically relevant variants, as well as the automated… Read more »
VarSeq Updated with CNV Annotations, CNV PhoRank, and Region Assessment Catalogs This year we have released multiple advances to support CNV analysis in VarSeq, expanding our target region based VS-CNV caller to handle exomes and low-depth genomes as well as additional supporting algorithms like calling Loss of Heterozygosity regions. To top this off, VarSeq 1.4.7 has been shipped with many… Read more »
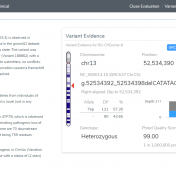
Variant interpretation is an integral part of any workflow that results in some decisions being made about the validity and suspected functional impact of a variant in a given sample and their presenting phenotypes. The VarSeq Assessment Catalog functionality is designed to assist the VarSeq user in streamlining this process. To include this functionality in your workflow, you will first… Read more »
Recently, a Golden Helix customer reached out to support for advice on how to validate the variant scoring between two different VarSeq users. Here we break down one possible solution as an opportunity to showcase the utility of both the Latest Sample Assessment function and an alternative way to leverage the Compute Fields function. To set up this validation project,… Read more »
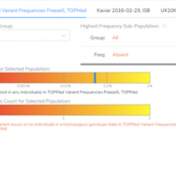
Our previous webcast demonstrated some of the new functionalities of VSClinical, including the ability to add ACMG frequency sources for the ACMG BA1, BS1, and PM2 criteria. This new feature was spurred by the feedback from our users, which requested supporting frequency tracks other than gnomAD Exomes and 1kG Phase3. Now, users can implement population catalogs to VSClinical such as… Read more »
It doesn’t take much effort to find articles discussing the value of Next-Generation Sequencing (NGS). There is a consistent tone amongst authors that implementing NGS pipelines are critical for clinical efficiency in both hereditary disorders and somatic. However, NGS strategies do not come without their own challenges. Challenges include not only the detection and calling of high quality/probability variants from… Read more »
With this two-part blog series, users should now be able to perform CNV analysis using their data, set up basic quality filter standards to isolate high-quality events and utilize annotations to hone in on publicly known events as well as in-house recorded CNVs from previous projects.
Precision oncology moves fast, and labs need tools that can keep up. Golden Helix CancerKB is built for clinical labs that want high-quality, report-ready cancer interpretations without the manual workload or inconsistencies that slow teams down. Stay in the Loop with Golden Helix CancerKB Oncology doesn’t stand still and neither does Golden Helix CancerKB. Our in-house curation team continually reviews… Read more »
VarSeq 3.0.0 is officially here, and it’s one of our most significant releases yet! The headline feature of this update is full integration with VSWarehouse 3, including a new login system that supports both single-user mode and server-managed authentication. Users can now choose to work locally or seamlessly store projects and data on a shared VSWarehouse location. There were several… Read more »
As Next-Generation Sequencing (NGS) becomes central to modern clinical diagnostics, laboratories are managing more data than ever before, and turning that data into actionable insight requires both precision and scalability. In our recent webcast, Next-Gen Sequencing at Scale: Transforming Per-Sample Outcomes into Institutional Knowledge with VSWarehouse, we explored how Golden Helix’s software ecosystem streamlines NGS analysis, bridging the gap between… Read more »
Clinical variant analysis experts with fully integrated workflows are often hesitant to upgrade or migrate their workspace, and rightly so. Similarly, those looking for new next-generation sequencing analysis software, whether for secondary (alignment and variant-calling) or tertiary (variant annotation, filtration, interpretation, and reporting) analysis, can be easily daunted by the breadth and depth of the process of setting up a… Read more »